5309
Age-Agnostic, Unsupervised Segmentation of Infant Brains using Magnetic Resonance Fingerprinting1Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 2Radiology, University of North Carolina, Chapel Hill, NC, United States
Synopsis
Keywords: Neuro, Segmentation, Atlas-Free
Brain segmentation is challenging in infants, as rapid changes to tissue properties and shapes during developmental growth make atlas-based modeling difficult. We use MRF-derived image features and density-based clustering to segment 2D brain slices from subjects without assumptions about subject age, brain shape, or image intensity. Segmentations from the proposed method closely match SPM without needing different atlases for subjects of varying ages. With flexible assumptions about the number of tissues present in an image, the proposed method identifies additional tissues such as CSF partial volume voxels and neonatal myelination that are not segmented by atlas-based approaches.Introduction
Neonatal brain segmentation faces several limitations1. MR contrast in neonates is approximately half that of adults, making it difficult to discern tissue boundaries. Supervised learning of segmentation requires tedious manual segmentation of ground truths2, and neonatal training data is difficult to acquire. Atlas-based segmentation3 uses atlases of expected brain shape and image intensity with assumptions about tissue types to impose segmentation expectations on input images. This technique struggles to generalize across infants, as T1- and T2-weighted contrasts and morphometry vary by month of age and infant atlases with one-month temporal resolution are unavailable. Atlas-based segmentation is also restricted to segmenting tissue types in the atlas and may incompletely classify partial volume voxels4 with multiple tissue types or unique tissues like myelin.Magnetic resonance fingerprinting (MRF) is a quantitative alternative to MRI that simultaneously measures T1 and T2 tissue properties in a single scan5. MRF quantification is independent of age or contrast, making it a prime candidate for age-agnostic segmentation. Rather than using MR contrast-weighted images with low CNR in baby images, MRF provides countless image contrasts via synthesis from signal evolutions or quantitative maps that highlight tissues and boundaries for segmentation. We propose a pipeline that selects MRF-based images to produce unsupervised tissue segmentations using a multi-image clustering approach.
Methods: MRF Image Features
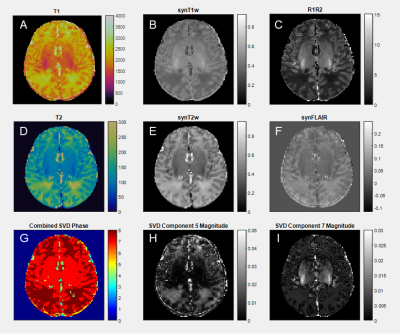
We use an unsupervised density-based clustering algorithm (HDBSCAN)6 to cluster selected image features and manual intervention to disqualify poor features. T1 and T2 maps are collected from MRF scans and used to synthesize clinical contrast-weighted images and quantitative maps. MRF signal evolutions are simulated voxel-wise per T1-T2 pair and SVD compressed7 to create signal component magnitude images. SVD phase images are binarized and combined to create a discretized image feature. Figure 1 shows the synthesized contrasts and best-performing features from a neonate MRF scan (10 days): T1, T2, synT1w, synT2w, synFLAIR, R1R2, combined phase of 8 SVD components, and magnitudes of SVD components 5 and 7.Methods: Segmentation Algorithm Design
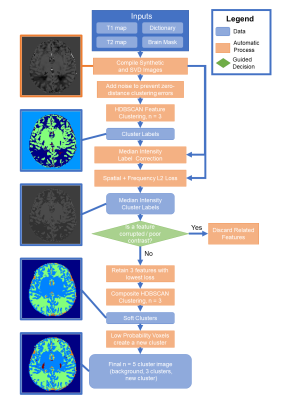
The proposed algorithm is summarized in Figure 2. Features are synthesized from MRF maps. ±0.5% of standard deviation uniformly distributed noise is added to each feature to prevent division by zero for distances between values. Each feature is HDBSCAN clustered to find a three-cluster solution. Clustering labels are intensity corrected to the median of the feature voxels of that label, creating a segmentation image that resembles the feature image.A combined image domain L2-norm and frequency domain magnitude L2-norm loss function is computed for each feature and its clustering to evaluate how each feature divides into distinct tissue types: $$L(f,l) = \lambda_1 \sum ||f - l||_2 + \lambda_2 \sum || |F| - |L| ||_2$$ where f is feature, l is intensity-corrected label, F and L are Fourier transforms of f and l, and λ are scalars. The user evaluates if any features are artifact corrupted. If any are deemed inappropriate, it and other features derived from it are discarded.
The three features with minimal L are compiled into a feature set and clustered again with HDBSCAN to produce a soft clustering. Voxels with low probability of belonging to any cluster are assigned to a fourth non-background cluster, which is observed to correspond to situationally unique tissues such as CSF partial volume voxels or infant myelination. This produces the final segmentation image without atlas or age assumptions.
Methods: Validation
Clustering was performed on 2D MRF slices from subjects of varying age (24-years-old, 10-day-old, 9-month-old) to investigate generalization across age. Reference clustering was performed on synthetic T1w using SPM8 with age-appropriate atlases9. DICE coefficients were calculated for grey matter, white matter, and CSF between the proposed method and SPM.Results
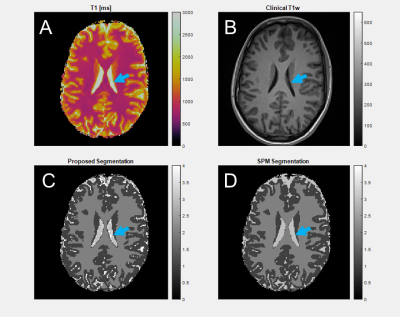
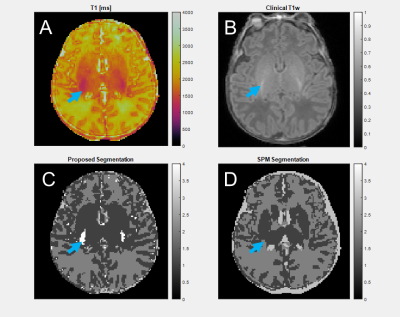
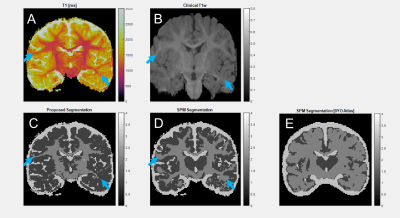
In a 24-year-old adult (Figure 3), grey matter DICE agreement was 0.9208, white matter 0.9502, and CSF 0.5375. Grey and white matter regions from the two methods are similar, but there is significant difference in CSF, where the proposed method identifies CSF partial volume voxels as distinct from the tissue types in the SPM atlas.In a 10-day-old neonate (Figure 4), the grey matter DICE score compared to SPM was 0.7005, white matter 0.7037, and CSF 0.4085. Grey and white matter segmentation is similar, though SPM segments the internal capsule via atlas assumptions despite the anatomy being poorly quantitatively differentiated. CSF is over-predicted by SPM relative to the proposed method. SPM requires a 0-year-old (0YO) atlas for segmentation, but the assumptions of our method do not change. An additional cluster is segmented around myelination in the basal ganglia, which is missed by SPM due to atlas limitations.
A 9-month-old subject (Figure 5) was SPM segmented with 0YO (Figure 5E) and 1YO (Figure 5D) atlases. This subject has reduced differentiation between grey and white matter due to incomplete contrast inversion. 0YO segmentation fails, while 1YO segmentation imperfectly segments white matter. For the more similar 1YO SPM segmentation compared to our method, DICE scores were 0.8072 for grey matter, 0.6958 white matter, and 0.8661 for CSF.
Conclusion
We present an age-agnostic tissue segmentation method using MRF. We report reasonable segmentation of grey and white matter without assumptions about contrast between tissue types. Future improvements include expanding to 3D segmentation, automating feature selection for a fully automatic algorithm, and incorporating spatial regularization for smoother segmentation maps.Acknowledgements
This work was supported by Siemens Healthineers and NIH grants R01 NS109439, R01 EB008374, and R01 EB006733.
References
1. Devi et al, Neonatal brain MRI segmentation: A review, Computers in Biology and Medicine, (2015) 64:163-178.
2. Singh and Singh, A Review of Publicly Available Automatic Brain Segmentation Methodologies, Machine Learning Models, Recent Advancements, and Their Comparison, Annals of Neurosciences, (2021) 28(1-2):82-93.
3. Balafar et al, Review of brain MRI image segmentation methods, Artif Intell Rev (2010) 33:261-274.
4. Deshmane et al, Partial volume mapping using magnetic resonance fingerprinting, NMR in Biomedicine, (2019) 32:e4082.
5. Ma et al, Magnetic Resonance Fingerprinting, Nature (2013) 495:187-192.
6. McInnes and Healy, Accelerated Hierarchical Density Based Clustering, 2017 IEEE International Conference on Data Mining Workshops, pp 33-42, 2017.
7. McGivney et al, SVD Compression for Magnetic Resonance Fingerprinting in the Time Domain, IEEE Trans on Med Imag (2014) 33:2311-2322.
8. Ashburner and Friston, Unified Segmentation, NeuroImage, (2005) 26:839-851.
9. Shi et al, Infant Brain Atlases from Neonates to 1- and 2-year-olds, PLoS ONE, (2011) 6:e18746.
Figures