5271
The Multi-Class Segmentation of the Human Cerebral Vasculature in TOF-MRA: A Supervised Deep Learning Approach1Departement of Biomedical Imaging and Radiation Science, Universite de Sherbrooke, Sherbrooke, QC, Canada, 2Departement of Computer Science, Universite de Sherbrooke, Sherbrooke, QC, Canada, 3Departement of Psychiatry, Harvard Medical School, Belmont, MA, United States, 4Departement of Radiology, Universite de Sherbrooke, Sherbrooke, QC, Canada
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Neuro, Software Tool
Currently, there is no accurate fully automated multi-class method to segment the whole cerebral arterial tree in time-of-flight magnetic resonance angiography (TOF-MRA). We developed an artificial intelligence based software tool to identify cerebral arteries in TOF-MRAs. We trained a neural network on a TOF-MRA dataset and labeled the cerebral arterial tree using different image processing techniques. Our software tool is fast and reliable, with no human intervention, and allows for the conduction of large-scale TOF-MRA studies while being versatile in segmenting a diverse set of TOF-MRAs.Introduction
Traditionally, the analysis of time-of-flight magnetic resonance angiography (TOF-MRA) is carried out via visual inspection or manual measurement. This process is neither scalable nor reliable, because it is time-consuming and prone to inter-reader variability. Currently, there is no fully automated method that can perform a multi-class segmentation of the whole cerebral arterial vasculature. We developed a fast, open-source and reliable artificial intelligence (AI) based solution that can accurately segment the whole cerebral arterial vasculature in a TOF-MRA, and classify different arteries using supervised machine learning.Methodology
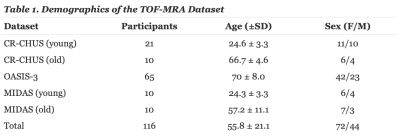
TOF-MRA DatasetThe TOF-MRA dataset consists of 116 MRAs that are diverse in fields of view, and scanner manufacturer. As shown in Table 1, 31 subjects were scanned locally at the research center in the Sherbrooke Fleurimont Hospital (CR-CHUS), 20 images were acquired from the MIDAS dataset and 65 from the Open Access Series of Imaging Study (OASIS-3) dataset. All MRAs were obtained from a 3T MRI scanner with resolutions of 0.51 × 0.51 × 0.80 mm³, 0.625 × 0.625 × 0.625 mm³ and 0.30 × 0.30 × 0.60 mm³ for MIDAS, CHUS and OASIS respectively. All subjects had no history of cerebrovascular disease.
Training Set
In each image in the training set, we used the Express IntraCranial Arteries Breakdown (eICAB1) to identify all major components of the Circle of Willis (CW). Next, a Sato2 filter was used to identify all voxels outside the CW belonging to a cerebral artery. A hysteresis3 threshold using threshold of 50% and 45% of the average sato score of basilar and the internal carotid arteries was applied, followed by a watershed4 algorithm to extend all CW labels into the distal vessel map. The approach ensured that all potential distal arteries connected back to the CW, and thus eliminated a large number of false-positives. An example of this approach is shown in Figure 1. All images were visually inspected prior to including them in the training stage.
The Training Stage
Training of the binary vessel images was carried out using the nnU-Net5 framework. The model was trained using the default parameters of a 1000 epochs, with a loss function consisting of the sum of dice and cross entropy loss, and validated with five-fold cross-validation.
Model Validation
To validate our model, we analyzed a series of new MRAs not included in the training set. To test its versatility, we analyzed 1 high-FOV 3T MRA and 1 7T MRA as shown in Figure 2. For accuracy, we analyzed 16 3T TOF-MRAs whose arterial vasculature were manually annotated by a human expert as shown in Figure 3. To test its consistency we evaluated its test-retest reliability using the Midnight6 dataset, a 10 subject dataset where each subject has 4 different TOF-MRA scans taken at different times.
Results
An important characteristic of our solution is its versatility and its ability to identify and segment TOF-MRAs from scanners with a variety of field strengths, resolutions and fields of view as illustrated in Figure 2. Additionally, it can handle different image artifacts.As a measure of accuracy the manually annotated TOFs were compared to our whole brain binary segmentation using dice coefficient and recall. The mean dice score and recall across the whole dataset is 71.28% and 72.73% respectively. To test our multi-class accuracy even further we calculated the dice score for independent artery groups such as the middle and anterior cerebral artery, yielding 68.97% and 68.87% respectively.
For the test-retest reliability, we compared one TOF-MRA scan with the 3 other scans of the same subject in MNI space, we attained a mean dice coefficient of 71.5% and recall of 71.64%. An example for our comparison is found in Figure 4. In contrast, a mean dice coefficient and recall of one TOF-MRA compared to 3 other scans from different individuals is 23.13% and 22.21% respectively.
Our method can segment a TOF-MRA image in around 10 minutes, which is a fraction of the time taken to fully segment the whole brain manually by hand.
Discussion
Taken together, our results demonstrate that our software tool yields an accurate and reliable whole brain arterial vessel segmentation without the need of any human involvement such as hand labeling or human corrections. In a matter of minutes, the trained model can generate a multi-class segmentation of the whole brain arterial vasculature of the input TOF image.Conclusion
Since accurate segmentations of TOF-MRAs are now possible, we look forward to having our software tool being used for localizing vascular abnormalities and calculating metrics such as an artery's diameter and volume. Thus, improving clinical practices and allowing for the conduction of large-scale TOF-MRA studies.Acknowledgements
I am grateful for the financial support I received through Universite de Sherbrooke's bourse d'excellence.References
- Dumais, F., Caceres, M. P., Janelle, F., Seifeldine, K., Arès-Bruneau, N., Gutierrez, J., Bocti, C., & Whittingstall, K. (2022, July 6). EICAB: A novel deep learning pipeline for circle of willis multiclass segmentation and analysis. NeuroImage. Retrieved November 3, 2022, from https://www.sciencedirect.com/science/article/pii/S1053811922005420
- Sato, Y., Nakajima, S., Atsumi, H., Koller, T., Gerig, G., Yoshida, S., & Kikinis, R. (1997, January 1). 3D multi-scale line filter for segmentation and visualization of curvilinear structures in medical images: Semantic scholar. semanticscholar.org. Retrieved November 3, 2022, from https://www.semanticscholar.org/paper/3D-Multi-scale-line-filter-for-segmentation-and-of-Sato-Nakajima/deadd4e6dfec0f5148cf888a4a5a9fd85d36e4e7
- Sornam, M., Kavitha, M., & Nivetha, M. (1970, January 1). Hysteresis thresholding based edge detectors for inscriptional image enhancement: Semantic scholar. undefined. Retrieved November 7, 2022, from https://www.semanticscholar.org/paper/Hysteresis-thresholding-based-edge-detectors-for-Sornam-Kavitha/ab4377b5009b28329b6e6c40025a4f0db3af91ca
- Parvati, K., Prakasa Rao, B. S., & Mariya Das, M. (2009, January 5). Image segmentation using gray-scale morphology and marker-controlled watershed transformation. Discrete Dynamics in Nature and Society. Retrieved November 7, 2022, from https://www.hindawi.com/journals/ddns/2008/384346/
- Isensee, F., Petersen, J., Klein, A., Zimmerer, D., Jaeger, P. F., Kohl, S., Wasserthal, J., Koehler, G., Norajitra, T., Wirkert, S., & Maier-Hein, K. H. (2018, September 27). NNU-net: Self-adapting framework for u-net-based medical image segmentation. arXiv.org. Retrieved November 3, 2022, from https://arxiv.org/abs/1809.10486
- Evan M. Gordon and Timothy O. Laumann and Adrian W. Gilmore and Dillan J. Newbold and Deanna J. Greene and Jeffrey J. Berg and Mario Ortega and Catherine Hoyt-Drazen and Caterina Gratton and Haoxin Sun and Jacqueline M. Hampton and Rebecca S. Coalson and Annie Nguyen and Kathleen B. McDermott and Joshua S. Shimony and Abraham Z. Snyder and Bradley L. Schlaggar and Steven E. Petersen and Steven M. Nelson and Nico U.F. Dosenbach (2020). The Midnight Scan Club (MSC) dataset. OpenNeuro. [Dataset] doi: 10.18112/openneuro.ds000224.v1.0.3
Figures
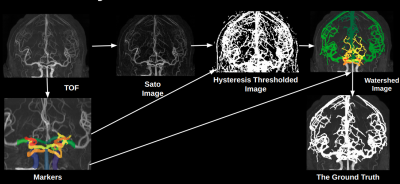
Figure1:
The summary of the process of ground truth labelling.
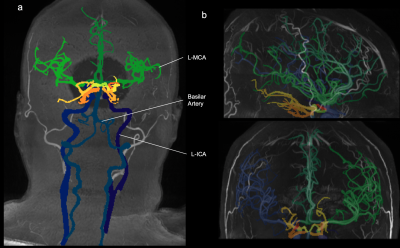
Figure 2:
a: Segmented TOF-MRA of 3T scanner
b: Segmented TOF-MRA from 7T scanner
All of the TOF-MRAs in the dataset are scanned with a 3T scanner, and their fields of view end at the top or center of the neckmuscles. This demonstrates our solutions versatility in handling TOF-MRAs with different fields of view and scanners’ fieldstrength that are not found in the training dataset.
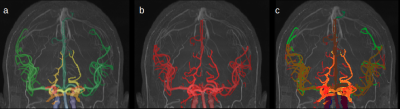
Figure 3:
a: The 18 label segmentation predicted by our solution
b: The hand corrected binary segmentation from the lab
c: An overlay of both segmentations, representing a dice score of 85.87%

Figure 4:
a: A binary segmentation of a patient’s TOF-MRA from the Midnight dataset registered in MNI space
b: A second segmentation of the same patient from a different time of the day overlapping segmentation a
c: A third segmentation of the same patient from a different time of the day overlapping the first 2 segmentations
d: A fourth segmentation of the same patient from a different time of the day overlapping the previous 3 segmentations
The mean dice coefficient between the 4 segmentations is 74.77%