5259
Towards a standardized file format for basis spectra and fitting results for linear-combination modeling
Helge J. Zöllner1,2, Kelley M. Swanberg3, John LaMaster4, Antonia Kaiser5, Jamie Near6, Candace Fleischer7,8, Brian J. Soher9, William T. Clarke10, and Georg Oeltzschner1,2
1The Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2Kennedy Krieger Institute, F. M. Kirby Research Center for Functional Brain Imaging, Baltimore, MD, United States, 3Department of Biomedical Engineering, Columbia University Fu Foundation School of Engineering and Applied Science, New York, NY, United States, 4Faculty of Computer Science, Technische Universität München, München, Germany, 5Department of Radiology and Nuclear Medicine, Amsterdam Neuroscience, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands, 6Physical Sciences Platform, Sunnybrook Research Institute, Toronto, ON, Canada, 7Department of Radiology and Imaging Sciences, Emory University School of Medicine, Atlanta, GA, United States, 8Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA, United States, 9Center for Advanced MR Development, Department of Radiology, Duke University Medical Center, Durham, NC, United States, 10Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom
1The Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2Kennedy Krieger Institute, F. M. Kirby Research Center for Functional Brain Imaging, Baltimore, MD, United States, 3Department of Biomedical Engineering, Columbia University Fu Foundation School of Engineering and Applied Science, New York, NY, United States, 4Faculty of Computer Science, Technische Universität München, München, Germany, 5Department of Radiology and Nuclear Medicine, Amsterdam Neuroscience, Amsterdam UMC, University of Amsterdam, Amsterdam, Netherlands, 6Physical Sciences Platform, Sunnybrook Research Institute, Toronto, ON, Canada, 7Department of Radiology and Imaging Sciences, Emory University School of Medicine, Atlanta, GA, United States, 8Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA, United States, 9Center for Advanced MR Development, Department of Radiology, Duke University Medical Center, Durham, NC, United States, 10Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom
Synopsis
Keywords: Data Analysis, Spectroscopy, open source
We propose a standardized format for storing basis sets and linear-combination modeling results. Conventions, guidelines, and conversion tools are currently developed in collaboration with the MRS community. This standardization effort will further improve data sharing and reproducibility, allowing for easier integration of MRS into larger neuroimaging frameworks.Introduction
Expert consensus recommends linear-combination modeling (LCM) for the quantification of virtually all in-vivo MRS experiments1. LCM models spectra as a linear combination of simulated metabolite basis spectra. The extracted information is only accurate if simulations are precisely reflecting the signal-acquisition experiment, i.e. use exact sequence timings and pulse and gradient waveforms2,3.Spectral simulation software packages generate outputs (basis spectra) in different high-level formats; either high-resolution free induction decays (FIDs) or transition table results that describe the signals in each FID as a collection of areas/phases/frequencies4–6. Users have to convert these to a low-level format that specific LCM software4,7–14 accepts, usually by down-sampling to the required digital resolution. While the LCModel basis set format (.basis) is widely used7,9, its capabilities of storing metadata are limited, e.g. for basis set creation provenance.
Similarly, no standardized format exists to store and communicate LCM fitting results. These include quantitative model parameters (concentrations and measurement uncertainties) and the overall fitted model4,7–14. This impedes data sharing, analysis provenance and integration into imaging and meta/mega-analysis frameworks15.
Recent community efforts towards standardization led to the development of NIfTI-MRS to supersede vendor-proprietary data formats16. NIfTI-MRS utilizes the NIfTI header-extension framework to store JavaScript Object Notation (JSON)-formatted metadata17. This allows bundling crucial documentation directly with the data.
Here, we introduce standardization guidelines for storing 1) high- and low-level basis spectra and 2) LCM fitting results. We describe open-source tools to convert among formats commonly used in MRS simulation and analysis packages. This will make basis sets and LCM fitting results accessible to a wider group of researchers to foster collaborative, reproducible, and transparent MRS research.
Methods
Figure 1 describes how the proposed new data formats integrate into a typical MRS workflow:- A spectral simulation tool generates a density-matrix representation (gray) of the metabolites based on a pulse sequence description, pulse sequence settings, and spin system descriptions.
- The density matrix representation is converted into the proposed high-level representation of the basis set (blue). JSON-formatted metadata include full provenance of the simulation process, e.g., software package, version, pulse timings/waveforms, and spatial grid settings. The basis set is either stored as high-resolution time-domain representations or as transition table values using generalized array representation readable by any programming language. A clear advantage of the transition table object is that it allows arbitrary resampling of the basis spectra.
- Conversion of the high-level representation to generate low-level basis spectra as input for the linear-combination algorithm. We propose to adopt NIfTI-MRS as a standardized format of the low-level representation (green); however, conversion to other third-party representations can be easily achieved.
- The low-level basis set is employed in linear-combination modeling, and the results are stored in NIfTI-MRS (orange, for fitted model).
We propose storage conventions for the NIfTI-MRS data container and its JSON-formatted metadata to hold information describing the low-level basis spectra and LCM results. Figure 2 shows their proposed NIfTI-MRS representations. Mandatory entries for basis sets in the NIfTI block are the time-domain data (dimension 4 ‘spectral time domain’) for different metabolites (dimension 6 ‘metabolites’). Flexible NIfTI-MRS dimension tags can optionally encode sub-experiments for e.g. edited spectroscopy (dimension 7 ‘editing’) or different metabolite moieties (dim 5 ‘moieties’), e.g. N-acetyl and aspartyl moieties from N-acetylaspartate. Simulation details and standardized metabolite naming conventions18 are stored in a simulation provenance header key.
For the LCM results, dimension 1-4 in the NIfTI-MRS array are reserved for spatial (dimension 1-3, optional) and spectral time domain (dimension 4, required) results. Higher dimensions (5-7) will store model components (mandatory; scaled basis functions, baseline terms, and residual) and dynamic results and sub-spectra for edited, time-resolved, or 2D data (optional).
Metadata will store descriptive names for each model component18, e.g., data, fit, residual, baseline, metabolite basis spectra. Optional entries may include modeling provenance (LCM cost function, regularization, priors, constraints, etc.). Model parameter storage (concentrations and uncertainties) will be developed alongside the above formats, either integrating with the proposed fitting results format or using appropriate stand-alone formats (such as NIfTI).
Results
A minimal example dataset to generate NIfTI-MRS representations of low-level basis spectra and LCM results using Osprey is freely available on GitHub (https://github.com/HJZollner/StandardBasisTools). We aim to introduce the proposed data formats into common analysis software (Figure 3). We will further develop scripts to convert fitting results from LCModel into NIfTI-MRS format (for MATLAB, these will be available in the same repository; for Python, they will be shared from the Vespa repository https://github.com/vespa-mrs).Discussion & Conclusion
We describe a standardized data format to store basis spectra and LCM results. This standard will: a) simplify basis set sharing; b) reduce the workload to support simulation-software-specific file formats; c) increase transparency and reproducibility by adding metadata and provenance; d) increase between-software compatibility; e) support MRS integration into existing software- and data-sharing frameworks. Development is spearheaded by the Code and Data Sharing Committee of the ISMRM MRS study group (forum.mrshub.org).Acknowledgements
This work has been supported by NIH grants R00 AG062230 and R21 EB033516.References
- Near J, Harris AD, Juchem C, et al. Preprocessing, analysis and quantification in single-voxel magnetic resonance spectroscopy: experts’ consensus recommendations. NMR Biomed. Published online February 21, 2020:e4257. doi:10.1002/nbm.4257
- Landheer K, Juchem C. Recommendations for Accurate Basis Set Generation for Magnetic Resonance Spectroscopy Quantification. In: 27th Annual Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). ; 2019.
- Gajdošík M, Landheer K, Swanberg KM, et al. The effect of basis sets on the analysis of in vivo brain MRS data obtained with standard PRESS sequences. In: 29th Annual Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). ; 2021.
- Soher BJ, Semanchuk P, Todd D, Steinberg J, Young K. VeSPA: Integrated applications for RF pulse design, spectral simulation and MRS data analysis. In: 19th Annual Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). ; 2011. Accessed May 19, 2020. https://cds.ismrm.org/protected/11MProceedings/files/1410.pdf
- Simpson R, Devenyi GA, Jezzard P, Hennessy TJ, Near J. Advanced processing and simulation of MRS data using the FID appliance (FID-A)—An open source, MATLAB-based toolkit. Magn Reson Med. 2015;77(1):23-33. doi:10.1002/mrm.26091
- Landheer K, Swanberg KM, Juchem C. Magnetic resonance Spectrum simulator (MARSS), a novel software package for fast and computationally efficient basis set simulation. NMR Biomed. 2021;34(5):e4129. doi:10.1002/nbm.4129
- Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30(6):672-679. doi:10.1002/mrm.1910300604
- Oeltzschner G, Zöllner HJ, Hui SCN, et al. Osprey: Open-source processing, reconstruction & estimation of magnetic resonance spectroscopy data. J Neurosci Methods. 2020;343:108827. doi:10.1016/j.jneumeth.2020.108827
- Wilson M, Reynolds G, Kauppinen RA, Arvanitis TN, Peet AC. A constrained least-squares approach to the automated quantitation of in vivo 1H magnetic resonance spectroscopy data. Magn Reson Med. 2011;65(1):1-12.
- Gajdošík M, Landheer K, Swanberg KM, Juchem C. INSPECTOR: free software for magnetic resonance spectroscopy data inspection, processing, simulation and analysis. Sci Rep. 2021;11(1):2094. doi:10.1038/s41598-021-81193-9
- Poullet JB, Sima DM, Simonetti AW, et al. An automated quantitation of short echo time MRS spectra in an open source software environment: AQSES. NMR Biomed. 2007;20(5):493-504. doi:10.1002/nbm.1112
- Graveron-Demilly D. Quantification in magnetic resonance spectroscopy based on semi-parametric approaches. Magn Reson Mater Phys Biol Med. 2014;27(2):113-130. doi:10.1007/s10334-013-0393-4
- Wilson M. Adaptive baseline fitting for MR spectroscopy analysis. Magn Reson Med. 2021;85(1):13-29. doi:10.1002/mrm.28385
- Clarke WT, Stagg CJ, Jbabdi S. FSL-MRS: An end-to-end spectroscopy analysis package. Magn Reson Med. 2021;85(6):2950-2964. doi:10.1002/mrm.28630
- Gorgolewski KJ, Auer T, Calhoun VD, et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci Data. 2016;3(1):160044. doi:10.1038/sdata.2016.441
- Clarke WT, Bell TK, Emir UE, et al. NIfTI-MRS: A standard data format for magnetic resonance spectroscopy. Magn Reson Med. 2022;88(6):2358-2370. doi:10.1002/mrm.29418
- Pezoa F, Reutter JL, Suarez F, Ugarte M, Vrgoč D. Foundations of JSON schema. In: Proceedings of the 25th International Conference on World Wide Web. ; 2016.
- Kreis R, Boer V, Choi IY, et al. Terminology and concepts for the characterization of in vivo MR spectroscopy methods and MR spectra: Background and experts’ consensus recommendations. NMR Biomed. 2021;34(5):e4347. doi:10.1002/nbm.4347
Figures
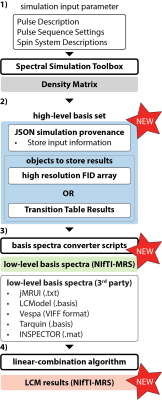
Figure 1 - LCM MRS analysis pipeline including the proposed standardized format: 1) Spectral simulation to generate density-matrix representations (gray), 2) conversion to new high-level basis set (blue), 3) conversion to low-level basis set, e.g., in the proposed NIfTI-MRS format (green) and 3rd-party formats as needed, and 4) linear-combination modeling and storage of final results in the proposed NIfTI-MRS format (orange).
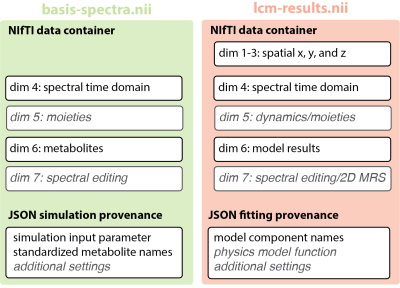
Figure 2 - Proposed NIfTI-MRS representation of the low-level basis spectra and LCM fitting results. Required and optional definitions are shown in black and gray italics, respectively.
Figure 3 - Spectral simulation software and MRS analysis packages that have agreed to support the new format at the time of submission.
DOI: https://doi.org/10.58530/2023/5259