5197
Glioma Classifications with 7T MR Spectroscopic Imaging
Sukrit Sharma1, Cornelius Cadrien2, Philipp Lazen1, Hangel Gilbert1, Roxane Licandro3, Wolfgang Bogner1, and Georg Widhalm2
1High-field MR Center, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, Medical University of Vienna, Vienna, Austria, 2Medical University of Vienna, Vienna, Austria, 3Department of Biomedical Imaging and Image-guided Therapy, Computational Imaging Research Lab (CIR) and Laboratory for Computational Neuroimaging, A.A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital / Harvard Medical School, Charlestown, MA, US., Medical University of Vienna, Vienna, Austria
1High-field MR Center, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, Medical University of Vienna, Vienna, Austria, 2Medical University of Vienna, Vienna, Austria, 3Department of Biomedical Imaging and Image-guided Therapy, Computational Imaging Research Lab (CIR) and Laboratory for Computational Neuroimaging, A.A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital / Harvard Medical School, Charlestown, MA, US., Medical University of Vienna, Vienna, Austria
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Brain, Glioma
To contribute to better tumour classification and thus enhancing patient outcomes, we statistically analysed metabolic maps of 37 glioma patients obtained using high resolution 7T MRSI. We tested and optimised different semi-supervised learning based classification approaches. Random forest classification of IDH mutation status and tumour grade in clinical imaging based segmented tumour regions yielded high diagnostic accuracy with AUC of 86% and 99% respectively. We found Glu, Gln, GSH, tCho, Ins, Gly and tCr as important determining features. These are similar to comparable SVS studies while providing the advantage of whole-brain coverage.Introduction
The WHO 2021 classification1 based tumour grading on molecular properties such as IDH-mutation, which could become powerful markers for determining tailored therapeutic approaches2 and thus patient outcomes. Current methods for tumour detection and classification come with both benefits and disadvantages. Cranial surgical biopsy and histopathological evaluations provide the most accurate diagnostic information while being highly invasive. Morphologic MRI cannot directly offer information on molecular and metabolic tumour properties. With 7T MRSI, we try to address these issues by creating a non-invasive method for molecular tumour classification based on the evaluation of altered tumour metabolism that is connected to tumour properties. In our previous research3, we saw trends in spectroscopic alterations in high-grade gliomas, such as increased Gln, Gly, and tCho and decreased NAA and tCr in tumour regions. Our current goal is to optimise statistical approaches for better accuracy in non-invasive tumour classifications.Methods
We recruited 36 glioma patients (median age: 52 ± 23) preoperatively (Figure 1). The 15-minute MRSI measurement was performed using a Siemens Magnetom 7T at a TR of 450ms, 64 x 64 x 39 matrix with isotropic 3.4 mm voxel3,4. In addition we acquired MP2RAGE and FLAIR with 0.8 mm isotropic resolution. The scans were post-processed using an in-house pipeline5 and quantified with LCModel6. An experienced neuroradiologist provided manual tumour segmentation based on routine 3T MRI. Segmentations included Necrosis (Nec), Contrast Enhancing (CE), and Non-Contrast Enhancing (NCE) regions. Metabolic ratio maps to total Creatine (tCr), total N- acetyl aspartate (tNAA), and total Choline (tCho) were calculated for further evaluations. We applied specific cutoff values based on past experience for each metabolite ratio map to limit the evaluation to the metabolic hotspot regions (no threshold, minimum threshold, median threshold, maximum threshold). We employed a Random Forest regression model and support vector machine (SVM) for grade and IDH mutation classification. For better statistical significance, we used Recursive feature elimination and cross-validation methods (leave-one-out). We plotted Receiver operating characteristic (ROC) curves were then plotted and calculated Area Under Curve (AUC) values to compare diagnostic accuracy of different evaluation models.Results
Depending on the applied metabolite thresholds and number of feature maps used, diagnostic accuracy for RF and SVM-based IDH mutation detection yielded AUC values ranging from 0.73-0.86 (Figure 2). Grade classification AUCs ranged from 0.71-0.99 (Figure 2). Highest AUC values were obtained with maximum threshold and using 3-6 selected features for the RF training model (Figure 3+4). We found Glu, Gln, GSH and tCho as the most important features in IDH classification. Ins, Gly, GSH and tCho were most crucial in grade evaluation. The best performing IDH mutation status classification model provided AUC of 0.86 with 6 features and max. threshold and AUC of 0.46 with tCho/tNAA as single feature and min. threshold (Figure 3). ROCs of the best performing tumour grade classification models showed AUC of 0.89 with 6 features and max. threshold. tCho/tNAA single feature AUC was 0.99 (FIgure 4). Manual evaluation of the spectroscopic maps and selected key features in 2 patients is shown in Figure 5.Conclusions
In this study, 7T MRSI yielded non-invasive glioma classification with high accuracy. The features that turned out to be the most important for IDH status determination and tumour grading were widely similar to comparable studies7 with larger cohort sizes. Our analysed patient sample is still too small to draw large-scale conclusions but we are still recruiting and expanding to pool MRSI data from multiple sites. The classification was based on manual tumour segmentation which could be automated in the future as well. The limitation of (3T) metabolic single-voxel-spectroscopy studies to rely on manual placement of the measured voxel does not exist with our whole-brain spectroscopic imaging sequence. Overall, we were able to show a promising application for 7T MRSI that points towards useful future insights into brain tumour metabolism, such as molecular profiling, delineation or treatment monitoring.Acknowledgements
This study was supported by the Austrian Science Fund (FWF) projects KLI-646 FW and KLI 1089-B as well as a 2021 Comprehensive Cancer Center grant of the Medical University of Vienna.References
1. Louis, D. N. et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro-Oncol. 23, 1231–1251 (2021).2. Han, S. et al. IDH mutation in glioma: molecular mechanisms and potential therapeutic targets. Br. J. Cancer 122, 1580–1589 (2020).3. Hangel, G. et al. High-resolution metabolic imaging of high-grade gliomas using 7T-CRT-FID-MRSI. NeuroImage Clin. 28, 102433 (2020).4. Hingerl, L. et al. Clinical High-Resolution 3D-MR Spectroscopic Imaging of the Human Brain at 7 T: Invest. Radiol. 55, 239–248 (2020).5. Považan, M. et al. Mapping of brain macromolecules and their use for spectral processing of 1 H-MRSI data with an ultra-short acquisition delay at 7 T. NeuroImage 121, 126–135 (2015).6. Provencher, S. W. Automatic quantitation of localizedin vivo1H spectra with LCModel. NMR Biomed. 14, 260–264 (2001).7. Ozturk‐Isik, E. et al. Identification of IDH and TERTp mutation status using 1 H‐MRS in 112 hemispheric diffuse gliomas. J. Magn. Reson. Imaging 51, 1799–1809 (2020).Figures
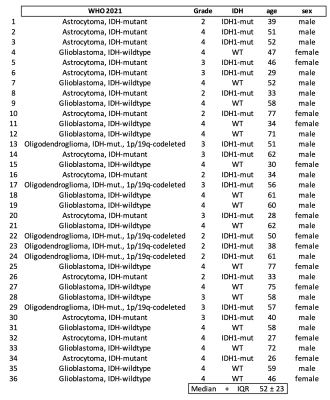
Fig 1: Patient overview according to guideline. Glioma classifications according to the latest WHO 2021 guidelines. IDH1 mutation status. Age and sex at time of measurement.
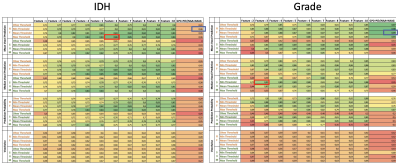
Fig 2: Comparison of random forest and SVM classifier results of sorted by applied thresholds. Multi feature with best performance is highlighted in red; tCho/tNAA single feature best performance highlighted in blue.
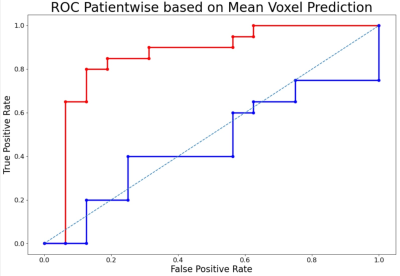
Fig 3: ROC of the best performing IDH mutation status classification models. Red: with 6 features and max. threshold (AUC = 0.86); Blue: tCho/tNAA as single feature and min. threshold (AUC = 0.46)
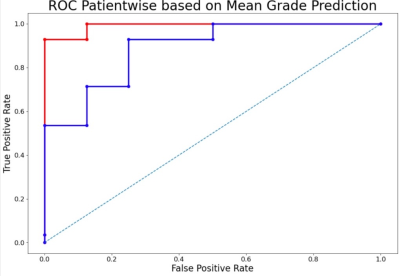
Fig 4: ROC of the best performing tumour grade classification models. Blue: with 6 features and min. threshold (AUC = 0.89); Red: with tCho/tNAA as feature and min. threshold (AUC = 0.99)

Fig 5: Spectroscopic and anatomic maps of 2 selected patients. Segmentation (green = non contrast enhancing tumour region; white = contrast enhancing tumour region). Top: Patient 2 (Astrocytoma, IDH mutant). Bottom: Patient 36 (Glioblastoma, IDH wildtype). Note the differences in spectroscopic ratio levels.
DOI: https://doi.org/10.58530/2023/5197