5194
Evaluating different k-space undersampling schemes with iterative and deep learning image reconstruction for fast multi-parameter mapping1Neurophysics, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 2Max Planck Computing and Data Facility, Garching (Munich), Germany, 3Neural Data Science and Statistical Computing, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 4Felix Bloch Institute for Solid State Physics, Faculty of Physics and Earth Sciences, Leipzig University, Leipzig, Germany
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction, Brain
Approaches for accelerating multi-echo gradient echo (ME-GRE) acquisitions as a basis for multi-parameter mapping (MPM) were explored. Fully sampled ME-GRE data were retrospectively undersampled to equispaced Cartesian, CAIPIRINHA and Poisson disc patterns. Echoes were jointly reconstructed with the iterative ENLIVE algorithm and the machine learning/artificial intelligence adapted DeepcomplexMRI (DCMRI) approach. The approaches result in comparable peak signal-to-noise ratio (PSNR) and structural similarity index measure (SSIM), but show different types and different levels of artifacts. The DCMRI approach promises fast reconstruction and flexibility in the choice of undersampling patterns for ME-GRE imaging in the future.Introduction
The well-established approaches to speeding up microstructural imaging using quantitative multi-parameter mapping (MPM)1,2 rely on Cartesian undersampling.3 Image reconstruction of the underlying multi-echo gradient echo (ME-GRE) data is performed echo by echo. Modern methods like compressive sensing (CS)4 have already been successfully applied to MPM5, but have not entered routine application nor addressed the multi-echo aspect of the data. This study explores different k-space undersampling schemes, i.e., elliptical Poisson Disc CS4. and CAIPIRINHA6, in comparison to equispaced Cartesian. The performance of iterative image reconstruction (ENLIVE)7 and machine learning based DeepcomplexMRI (DCMRI)8 on these datasets is evaluated.Methods
Fully sampled, 1mm isotropic resolution MPM datasets (a pair of PD- and T1-weighted 3D ME-GRE acquisitions with 8 equidistant echoes) were acquired on eleven healthy volunteers employing prospective motion correction at 3T (Connectom, Siemens Healthcare, Erlangen, Germany) with a 32-channel RF receive head coil.2,9 Fourier transform was applied in the readout direction, resulting in a stack of 2D k-space planes/slices. The resulting 2D k-space planes for each volume were processed separately, retrospectively undersampled and fed into the different reconstruction algorithms using all 8 echoes stacked together. The resulting images were combined with root sum of squares across RF coils and stacked back into a 3D volume. The dataset of one volunteer was held back for evaluation. Results were compared against root sum of squares coil combined fully sampled reference data in terms of peak signal-to-noise ratio (PSNR), structural similarity metrics (SSIM)10 and qualitative appearance. Voxel intensities of ENLIVE reconstructions were linearly scaled to best match the fully sampled reference.ENLIVE was applied using default parameters with 12 iterations in about 40 minutes per volume on 128 compute cores when processing 32 planes in parallel using 4 cores each. DCMRI was adapted to process all 8 echoes simultaneously by introducing an additional input/output dimension and modifying the necessary data preparation steps. The residual blocks were adjusted to use convolutions with 64 output channels for the hidden states. The training was performed separately for each sampling pattern employing an accelerated High-Performance-Computing node with 4x NVIDIA A100 GPU (40 GB HBM2 memory) for about 12 hours each. Inference for each volume was performed in less than 4 minutes on a single GPU.
Results
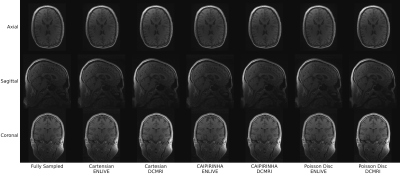
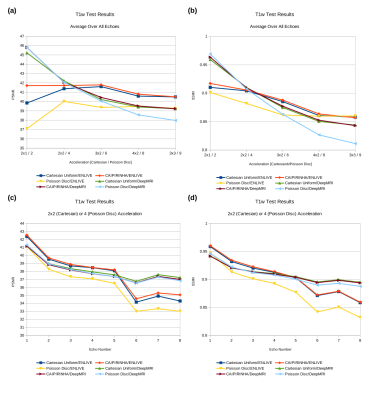
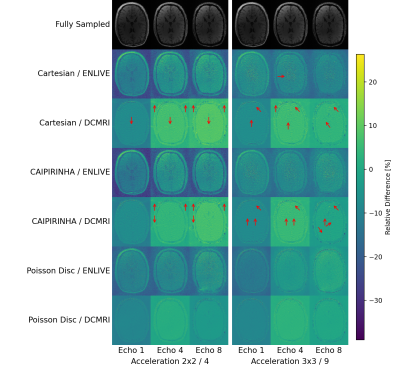
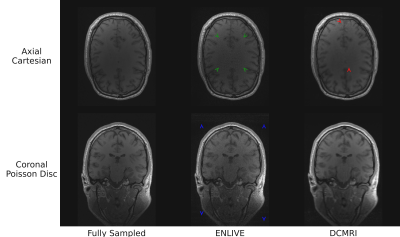
In general, image quality was rather high even at high acceleration factors (Fig. 1). Average PSNR and SSIM were comparable between k-space schemes and reconstruction methods, with PSNR= 36.4-45.8 dB and SSIM= 0.80-0.97. Higher acceleration resulted in reduced PSNR and SSIM (Figs. 2a and 2b). DCMRI yielded higher SSIM and PSNR than ENLIVE at lower acceleration but showed a steeper decrease in SSIM with increasing acceleration than ENLIVE. Independent of the method a similar quality reduction was found for longer echo times (Figs. 2c and 2d). ENLIVE showed a particular drop in quality for echo 6 and later echo times, which was not observed for DCMRI. Visual inspection shows that Cartesian and CAIPIRINHA sampling schemes resulted in more structured artifacts specifically when using DCMRI reconstruction (Fig. 3). ENLIVE tends to enhance noise and shift the average image intensity for slices with little signal (inferior and superior parts of the volume) leading to visible stripe like structures at the top and bottom in the coronal and sagittal views (Fig. 1 and Fig. 4).Discussion
When analyzing PSNR and SSIM a steeper decrease with higher acceleration factors for DCMRI is noticeable. One potential explanation is little robustness of the DCMRI architecture to very noisy data that might be handled by introducing batch normalization or dropout layers and potentially focusing stronger on image foregrounds during training. For ENLIVE, the sudden drop after echo 6 in PSNR and SSIM is unexpected. More stable results might be possible with further optimization of the hyper-parameters and regularization method used for the reconstruction. Also, the difference between average voxel intensities of ENLIVE reconstructions and fully sampled images should be further investigated. While ENLIVE can be used on almost any data with just hyper-parameter tuning, DCMRI has to be retrained for changed input data. This makes ENLIVE more tractable for sequences under development undergoing regular changes. However, due to long computation times, ENLIVE requires specialized hardware or offline reconstruction. On the other hand, the DCMRI reconstruction can be performed very fast with standard hardware. This gives DCMRI a clear advantage for established scanning protocols and allows for online reconstruction. In addition, DCMRI may be extended to include further processing steps like estimation of quantitative parameters in the future.Conclusion and Outlook
We compared state-of-the-art ENLIVE to multi-echo DCMRI, which showed high image quality even at 9x acceleration. The much shorter reconstruction time of DCMRI promises to facilitate routine high throughput applications and online visual image inspection. Future work will concentrate on establishing advantages of joint multi-echo DCMRI and ENLIVE reconstruction for calculation of MPMs and applying DCMRI to high-resolution data that suffers from long reconstruction runtimes.Acknowledgements
The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013) / ERC grant agreement n° 616905.
This project has received funding from the Federal Ministry of Education and Research (BMBF) under support code 01ED2210.
NW has received funding from the European Union's Horizon 2020 research and innovation programme under the grant agreement No 681094, and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – project no. 347592254 (WE 5046/4-2 and/or KI 1337/2-2)
References
1. Weiskopf N, Suckling J, Williams G, et al. Quantitative multi-parameter mapping of R1, PD*, MT, and R2* at 3T: a multi-center validation. Front Neurosci. 2013;7:95.
2. Pine KJ, Edwards LE, Callaghan MF, et al. Ultra-high resolution in vivo multi-parameter mapping of the human brain. in Proc Intl Soc Mag Reson Med 25. 2017:1168.
3. Leutritz T, Seif M, Helms G, et al. Multiparameter mapping of relaxation (R1, R2*), proton density and magnetization transfer saturation at 3 T: A multicenter dual-vendor reproducibility and repeatability study. Human Brain Mapping. 2020;41(15):4232–4247.
4. Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182–1195.
5. Berg R, Leutritz T, Weiskopf N, and Preibisch C. Multi‐parameter quantitative mapping of R1, R2*, PD, and MTsat is reproducible when accelerated with Compressed SENSE. Neuroimage. 2022;253:119092.
6. Breuer FA, Blaimer M, Heidemann RM, et al. Controlled aliasing in parallel imaging results in higher acceleration (CAIPIRINHA) for multi-slice imaging. Magn Reson Med. 2005;53(3):684–691.
7. Holme HCM, Rosenzweig S, Ong F, et al. ENLIVE: An Efficient Nonlinear Method for Calibrationless and Robust Parallel Imaging. Sci Rep. 2019;9(1):1–13.
8. Wang S, Cheng H, Ying L, et al. DeepcomplexMRI: Exploiting deep residual network for fast parallel MR imaging with complex convolution. Magn Reson Imaging. 2020;68:136–147.
9. Vaculčiaková L, Podranski K, Edwards LJ, et al. Combining navigator and optical prospective motion correction for high-quality 500 μm resolution quantitative multi-parameter mapping at 7T. Magn Reson Med. 2022;88(2):787–801.
10. Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image Quality Assessment: From Error Visibility to Structural Similarity. IEEE Trans Image Process. 2004;13(4):600–612.
Figures