5189
8-minute Rapid Whole-brain Diffusion Spectral Imaging with Deep Learning-based Reconstruction: A Feasibility Study1GE Healthcare MR Research, Beijing, China, 2Department of Radiology, Xuanwu Hospital, Capital Medical University, Beijing, China
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence
This study aims to shorten the diffusion spectral imaging (DSI) scan time to a clinically acceptable level while providing satisfactory complex white matter fiber structure description as well as accurate diffusion metric quantification by applying deep learning-based reconstruction. Images were acquired using conventional (≈30 min) and rapid (≈8 min) DSI sequences, and reconstructed using conventional and DL-based methods, respectively. Atlas-based fiber-tracking and diffusion metrics quantification from various advanced models were conducted. The results demonstrated that the 8-minute rapid DSI sequence combined with DL-recon can provide complex fiber structure tractography and advanced diffusion metric quantification of satisfactory quality.
Introduction
Diffusion spectral imaging (DSI) is of great value in neuroscience as it can accurately describe the microstructure changes of brain white matter and resolve the crossing fibers1,2. Also the data set acquired with DSI strategy enables the computation of a variety of advanced diffusion quantitative models, such as diffusional kurtosis imaging (DKI)3, neurite orientation dispersion and density imaging (NODDI)4, free water diffusion tensor imaging (FW-DTI)5, and mean apparent propagator (MAP)6. However, the practical use of conventional DSI is hurdled by the scan time that is generally over 30 minutes. AIRTM recon DL, a novel deep learning (DL) based MR reconstruction method, was demonstrated to be beneficial for diffusion image reconstruction and white matter fiber tractography by improving image signal-to-noise ratio (SNR) and sharpness, and reducing Gibbs-ringing artifacts7-9. This study aims to combine DSI scan with DL-based reconstruction to shorten the scan time to a clinically acceptable level while providing satisfactory crossing fiber resolving results and accurate diffusion metric quantification.Methods
Data acquisition & reconstruction 5 health volunteers were recruited and scanned on a 3T MRI scanner (SIGNATM Premier, GE Healthcare, Waukesha, WI) using a 48-channel head coil after obtaining written informed consent. Each scan contained two different DSI sequences. For the first sequence (conventional DSI): FOV = 22.4×22.4 cm2, voxel size = 2 mm isotropic, 75 slices without gap were collected to achieve whole-brain coverage, ARC acceleration factor = 2 and hyperband factor = 2, TR/TE = 6708/70 ms, 258 diffusion encoding directions with bmax = 3000 s/mm2 were acquired in 29min4s. The scan parameters of the second sequence (rapid DSI) were almost the same with conventional DSI except hyperband factor = 3, TR = 3738 ms, number of diffusion encoding directions = 128, and scan time = 8min6s. The rapid DSI raw data were reconstructed twice using conventional and DL-based reconstruction (Gibbs-ringing removal and 75% denoising), respectively. Therefore, for each subject, three sets of DSI images were collected: conventional DSI, rapid DSL, and rapid DSI with DL-recon.Data post-processing The DSI datasets were firstly pre-processed (including head-motion correction, eddy-current induced distortion correction, and skull-stripping) using FSL10 and MRtrix311. Subsequently, atlas-based deterministic fiber-tracking focusing on fiber-crossing regions was performed using DSI studio (http://dsi-studio.labsolver.org). Meanwhile, the computations of various diffusion metrics from the aforementioned models (DKI, NODDI, FW-DTI, and MAP) were performed using the DIPY12 and DMIPY13 toolboxes. The tractography and quantification results of three DSI datasets of each subject were compared to evaluate the efficacy of DL-recon.
Results
As shown in Figure 1, the rapid DSI sequence can drastically shorten the scan time by using a higher hyperband acceleration factor and fewer diffusion encoding directions, at the cost of extremely low SNR of diffusion images and quantitative metric maps under conventional reconstruction. The application of DL-recon significantly improved the SNR and quality of all images and maps. The tractography results for complex white matter fiber structures (Figure 2) manifest that the rapid DSI lost the ability to resolve crossing fibers, which can be restored by implementing DL-recon. Figure 3 demonstrates that the rapid DSI with DL-recon can provide abundant quantitative diffusion metrics with similar quantitative accuracy to conventional DSI only with a fraction of the data set and hence the scan time. Moreover, it is worth noting that the obvious Gibbs-ringing artifacts, which may be due to the relatively low in-plane resolution of the DSI scan and will theoretically contaminate both conventional and rapid DSI quantitative maps, can be effectively eliminated by DL-recon. From this perspective, rapid DSI with DL-recon can even provide higher quality quantitative results than conventional DSI.Discussion
The propagation of DSI to clinical practice is hurdled by its long scan time, and rapid DSI fits within the clinical scan frame but suffers from inferior SNR and hence degraded accuracy. In this work we investigated the use of deep learning-based reconstruction (AIRTM Recon DL) to improve the SNR of an 8-minute rapid DSI prototype. The DL-recon based rapid DSI showed similar efficacy to the 30-minute conventional DSI sequence in terms of complex fiber bundle structure analysis and advanced diffusion metric quantification. This mainly benefitted from the substantial improvement in image SNR by DL-recon, which can compensate for the SNR loss caused by bold acceleration/under-sampling in either k- or q-space. Furthermore, the DL-recon based rapid DSI may improve quantification accuracy compared to conventional DSI by eliminating Gibbs-ringing artifacts. Acquiring DSI images with satisfactory quality in acceptable scan time can greatly improve the clinical application value of this technique and effectively reduce the risk of artifact contamination caused by head motion.Conclusion
The proposed 8-minute rapid DSI sequence combined with DL-recon can provide whole-brain images, complex white matter fiber tractography, and advanced diffusion metric quantification of satisfactory quality.Acknowledgements
No acknowledgement found.References
[1] Wedeen V J, Hagmann P, Tseng W Y I, et al. Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging[J]. Magnetic resonance in medicine, 2005, 54(6): 1377-1386.
[2] Wedeen V J, Wang R P, Schmahmann J D, et al. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers[J]. Neuroimage, 2008, 41(4): 1267-1277.
[3] Jensen J H, Helpern J A, Ramani A, et al. Diffusional kurtosis imaging: the quantification of non‐gaussian water diffusion by means of magnetic resonance imaging[J]. Magnetic Resonance in Medicine, 2005, 53(6): 1432-1440.
[4] Zhang H, Schneider T, Wheeler-Kingshott C A, et al. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain[J]. Neuroimage, 2012, 61(4): 1000-1016.
[5] Pasternak O, Sochen N, Gur Y, et al. Free water elimination and mapping from diffusion MRI[J]. Magnetic Resonance in Medicine, 2009, 62(3): 717-730.
[6] Özarslan E, Koay C G, Shepherd T M, et al. Mean apparent propagator (MAP) MRI: a novel diffusion imaging method for mapping tissue microstructure[J]. NeuroImage, 2013, 78: 16-32.
[7] Wang X, Ersoz A, Litwiller D, et al. Robust Diffusion-Weighted Imaging with Deep Learning-Based DW PROPELLER Reconstruction. Abstract No. 3919, ISMRM, London, 2022.
[8] Choi K S, Figee M, Lebel R M, et al. Evaluation of the efficacy of a Deep Learning-based Reconstruction in the Connectomic Deep Brain Stimulation. Abstract No. 3357, ISMRM, London, 2022.
[9] Wang X, Yang B, Label M R, et al. High-resolution Diffusion Tensor Imaging at 7T with Multi-band Multi-shot EPI acquisition and Deep Learning Reconstruction. Abstract No. 3966, ISMRM, London, 2022.
[10] Jenkinson M, Beckmann C F, Behrens T E J, et al. Fsl[J]. Neuroimage, 2012, 62(2): 782-790.
[11] Tournier J D, Smith R, Raffelt D, et al. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation[J]. Neuroimage, 2019, 202: 116137.
[12] Garyfallidis E, Brett M, Amirbekian B, et al. Dipy, a library for the analysis of diffusion MRI data[J]. Frontiers in neuroinformatics, 2014, 8: 8.
[13] Fick R H J, Wassermann D, Deriche R. The dmipy toolbox: Diffusion mri multi-compartment modeling and microstructure recovery made easy[J]. Frontiers in neuroinformatics, 2019, 13: 64.
Figures
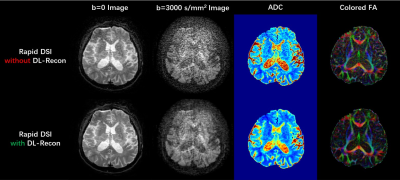
Fig. 1. Images (and corresponding quantitative metric maps) acquired using rapid DSI sequence in 8 min, and reconstructed using conventional (upper row) and DL-based method (lower row). ADC: apparent diffusion coefficient; FA: fractional anisotropy.
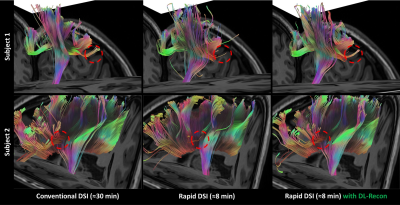
Fig. 2. Atlas-based white matter fiber tractography results of different DSI datasets (focusing on fiber-crossing regions). The tractography results of rapid DSI cannot resolve the association fiber derived from the cross region of the left corticospinal tract and the superior longitudinal tract (upper row) or the frontal aslant tract derived from the cross region of the left corticospinal tract and the corpus callosum body (lower row). On the contrary, the rapid DSI with DL-recon dataset showed similar crossing fiber resolving ability to conventional DSI dataset.
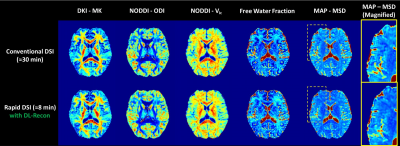
Fig. 3. Quantification results of various key metrics from multiple advanced diffusion models using different DSI datasets. The yellow dotted block regions in MAP-MSD maps were magnified and shown in the rightmost column to reflect the Gibbs-artifact elimination efficacy of DL-recon. MK: mean kurtosis; ODI: orientation dispersion index; Vic: intra-cell volume fraction; MSD: mean squared displacement.