5187
Myocardial strain generation from cine MR images using an automated deep learning network1Biomedical Engineering, Medical College of Wisconsin, Milwaukee, WI, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Heart
Current gold-standard method for obtaining myocardial strain is based on MRI tagged images, although this increases the MRI exam time and requires special analysis software. We propose to use cine MR images to train a deep neural-network to generate myocardial strain based on target strain maps generated from tagged images acquired at the same locations and timepoints as the cine images. The results showed high agreement between the output and target strain maps. Our method not only saves MRI scan time by acquiring only cine images but also pre and post image processing time by quantifying myocardial strains automatically.Synopsis
Current gold-standard method for obtaining myocardial strain is based on MRI tagged images, although this increases the MRI exam time and requires special analysis software. We propose to use cine MR images to train a deep neural-network to generate myocardial strain based on target strain maps generated from tagged images acquired at the same locations and timepoints as the cine images. The results showed high agreement between the output and target strain maps. Our method not only saves MRI scan time by acquiring only cine images but also pre and post image processing time by quantifying myocardial strains automatically.Introduction
Tagged MRI images are used as a current gold-standard technique to evaluate regional cardiac function1. Although MRI feature tracking allows for quantification of myocardial contractility patterns2, 3, the cine images lack the intramyocardial markers, which makes MRI feature tracking more suitable for global strain analysis. Nevertheless, acquiring both cine and tagging acquisitions results in longer the scan time. Therefore, attempts to reduce scan time while obtaining both global and regional cardiac function parameters would be appreciated. With recent development in deep learning (DL) capabilities, we sought to develop a DL-based network that can generate myocardial strain maps from conventional cine images after training the network with cine images and matched strain maps generated from corresponding tagged images acquired at the same locations and timepoints of the cine images.Methods
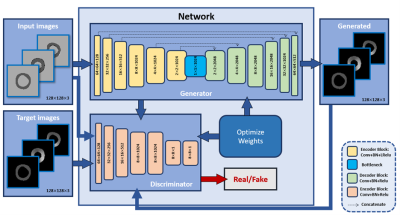
The developed framework is based on the image-to-image translation approach,4 which is built upon two main models, generator and discriminator. Our fine-tuned architecture (Figure 1) of the generator has 7 convolutional layers, each of which contains convolution, batch normalization (BN), and leaky ReLu activation during the network contraction path. The expansion path then follows, which includes 7 deconvolutional stages, each of which contains deconvolution, batch normalization, and ReLu activation, and then ends with a tanh activation function. The discriminator includes 4 stages of convolution, batch normalization, ReLu with kernel size of (4,4), and ends with a sigmoid function. The inputs of the network are cine difference images, generated by subtracting consecutive cine images to extract the endo- and epicardium motion while the targets of the network are corresponding myocardial strain maps in the radial and circumferential directions generated from the tagged images, which are acquired at the same slices and cardiac phases as the cine images, using the SinMod method5. Both cine and tagged images are automatically segmented by Unet network6. The datasets included a total of 1134 images of basal, mid-ventricular, and apical short-axis slices. The developed network was trained on 907 images and tested on 227 images to generate radial and circumferential strains.Results
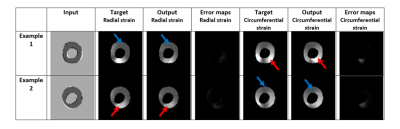
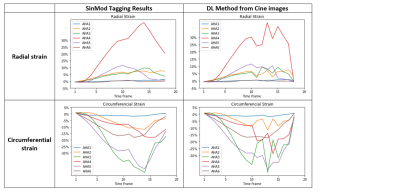
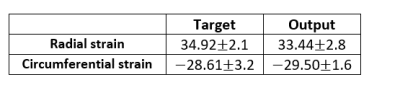
The generated output radial and circumferential strains (Figure 2, Table 1) showed myocardium shape similar to that in the input images as well as regional bright and dark signal intensities (representing tissue strains) at corresponding locations to those in the gold-standard myocardial strains generated from the tagged images. The dice scores were 0.97 in both radial and circumferential strains. The mean standard deviation of radial strains are 34.92 2.1 and 33.44 2.8 from target and output images, respectively and of circumferential strains are -28.61 3.2 and -29.50 1.6 from target and output images, respectively. Figure 3 shows strain curves at different time points generated using gold-standard analysis of the tagged images versus the corresponding curves generated using the proposed DL technique, which shows good agreement between the results from the two approaches. Student t-test showed insignificant strain measurement differences between all paired strain measurements (p>0.05). Lin’s concordance correlation coefficients (CCC) were 0.92 and 0.87 for radial and circumferential strains, respectively, reflecting that high agreement between the target and the output values. The developed method reduced the time required for generating the myocardial strains by two orders of magnitude to a sub-second without the need for pre- or post-processing.Discussion and conclusion
The developed DL-based neural network generates myocardial strains directly from the cine images with good agreement with results from gold standard tagged images. Therefore, the proposed network enables automatic quantification of myocardial strain without the need for tagged images. Potentially, this network can be used to reduce MRI scan time and allow for more adoption of strain imaging for evaluating different cardiovascular diseases.Acknowledgements
This work was supported by Daniel M. Soref Charitable Trust, MCW, USA.References
1. Ibrahim EH, Stojanovska J, Hassanein A, et al. Regional cardiac function analysis from tagged MRI images. Comparison of techniques: Harmonic-Phase (HARP) versus Sinusoidal-Modeling (SinMod) analysis. Magn Reson Imaging. 2018;54:271-282.
2. van Everdingen WM, Zweerink A, Nijveldt R, et al. Comparison of strain imaging techniques in CRT candidates: CMR tagging, CMR feature tracking and speckle tracking echocardiography. The international journal of cardiovascular imaging. 2018;34:443-456.
3. Schuster A, Hor KN, Kowallick JT, Beerbaum P, Kutty S. Cardiovascular Magnetic Resonance Myocardial Feature Tracking: Concepts and Clinical Applications. Circ Cardiovasc Imaging. 2016;9:e004077.
4. Isola P, Zhu J-Y, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. Proceedings of the IEEE conference on computer vision and pattern recognition2017:1125-1134.
5. Arts T, Prinzen FW, Delhaas T, Milles JR, Rossi AC, Clarysse P. Mapping displacement and deformation of the heart with local sine-wave modeling. IEEE Trans Med Imaging. 2010;29:1114-1123. 6. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. 2015.
Figures