5186
DeepMRS-Net: QUANTIFICATION OF MAGNETIC RESONANCE SPECTROSCOPY MEGA-PRESS DATA USING DEEP LEARNING1Biomedical Engineering, Columbia University, New York, NY, United States, 2Columbia University, New York, NY, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Brain, Convolutional Neural Networks, Multi-class Regression, Unsupervised Learning
Quantification of metabolites in the human brain in vivo from magnetic resonance spectra (MRS) has many applications in medicine and psychology, but it remains a challenging task despite considerable research efforts. In this paper, we propose quantification of metabolites from MEGA-PRESS data using deep learning through an unsupervised learning approach. A regression framework based on the Convolutional Neural Networks (CNN) is introduced for estimation of spectral parameters including the relative concentrations of metabolites, line-broadening, and zero-order phase. The results show that the model is capable of reliably fitting in vivo data.Introduction
Traditional MRS quantification methods involve solving an optimization problem of minimizing the difference between the target spectrum and a given parameterized model function. The parameters are usually estimated by fitting the model using a known basis set of the metabolite signals. Despite numerous proposed fitting methods, an accurate and reliable way of quantifying in vivo brain metabolites remains difficult [1].As machine learning becomes more prominent in the field of medical imaging, Chandler et. al [2] applied deep learning for MRS quantification. Their model, named MRSNet, is a CNN that operates on five metabolites. While MRSNet has shown promise on labeled simulated data and phantom data, it has not been applied to in vivo brain data for which there is no ground truth. On the other hand, our approach is based on unsupervised learning where the model attempts to minimize the residual between the predicted spectra and input target spectra, in a way that is reminiscent of the traditional fitting methods. This allows our model to perform quantification on both simulated and in vivo data alike without the need for ground truth data.
Methods
2.1 Data2.1.1 In vivo Data
Data from several 3T MRI scanners from three major manufacturers (GE, Philips, Siemens) are obtained from the Big GABA [3] repository. Of which, the subset of data from Phillips scanners are used in this study, for a total of 101 pairs of edit-ON/-OFF spectra with the following common parameters: TR/TE = 2000/68 ms, 320 averages, editing at 1.9/7.46 ppm for edit-ON/-OFF, respectively.
2.1.2 Simulated Data
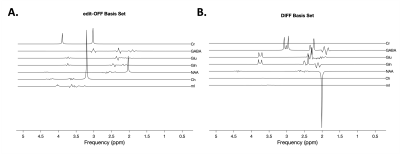
A basis set containing seven metabolites of interest is simulated using Osprey [4] using the same scanning parameters as those of the in vivo data.The relative concentrations of the individual metabolites for the simulated spectra are sampled by a Sobol sequence. The concentrations are used to produce a linear combination of the basis set. Figure 2 shows the basis sets and a few examples of the simulated spectra.2.1.3 Data Pre-processingFor the in vivo data, the individual transients are processed using the CNN-SR model proposed by Ma et. al [5] for frequency and phase correction before they are averaged. For both the simulated and in vivo data, the imaginary component is discarded, and the frequency-domain spectra are trimmed to obtain a length of 350 in the range from 4.2 ppm to 1.5 ppm. Finally, the trimmed spectra are normalized so that the largest peak has an amplitude of ± 1.
2.2 Network Architecture
The input to DeepMRS-Net is a 350 x 2 matrix, where the row represents the data points in the spectrum and the columns represent the OFF and DIFF spectra. The model consists of five 1D convolutional layers, each followed by a 1D max-pooling layer. The final fully-connected layer outputs 9 values corresponding to the predicted values of the seven concentrations, the line-broadening time constant, and the zero-order phase angle. To make unsupervised learning possible, extra tensor operations are appended to the end of the output layer so that a predicted spectra can be generated as shown in Fig 1. The final predicted spectra have the same dimension and channel size as the input. The loss function is simply the MAE between the output predicted spectra and the input target spectra.
Results
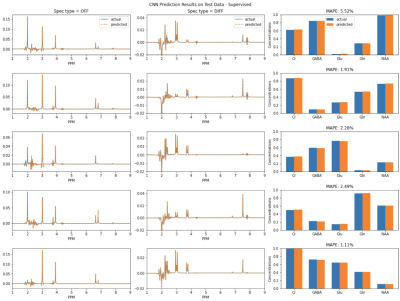
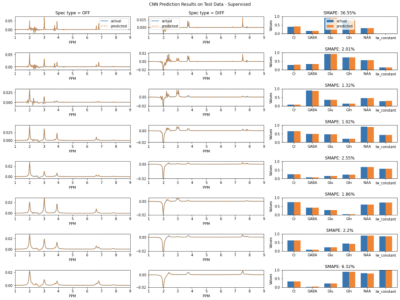
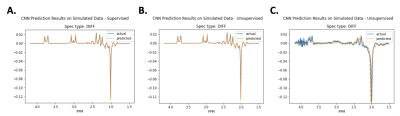
Figure 3 shows the prediction results on simulated data using supervised learning. Figure 4 shows the prediction results on simulated data using unsupervised learning. Figure 5 shows results for in vivo data.Discussion
Importantly, training the model on simulated data helps to confirm the assumption that minimizing spectral loss during unsupervised learning leads to accurate quantification of the metabolites. Due to the overlapping signals of several metabolites, a different linear combination of the metabolite basis signals could have the same spectral appearance as the target, but different underlying concentrations. Since the average MAPE between the predicted values and the labels for 20,000 simulated spectra is quite low, the results indicate that a good fitting of the target spectra could reliably lead to accurate estimations of the metabolite concentrations.Conclusion
Quantification of metabolites in MRS imaging using an unsupervised learning approach is presented for the first time. Training the model via unsupervised learning using spectral loss has an advantage in that it requires no ground truth. In addition, a common downside of using machine learning is that efficient training of the models usually requires a large number of samples and such data is not easily available in vivo. Our model circumvents this challenge as it is based partly on traditional fitting approaches. DeepMRS-net shows promise in working with both simulated data and in vivo data.Acknowledgements
No acknowledgement found.References
1. Hatami, N., Sdika, M., Ratiney, H. (2018). Magnetic Resonance Spectroscopy Quantification Using Deep Learning. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11070. Springer, Cham. https://doi.org/10.1007/978-3-030-00928-1_53
2. Chandler, M., Jenkins, C., Shermer, S., & Langbein, F.C. (2019). MRSNet: Metabolite Quantification from Edited Magnetic Resonance Spectra With Convolutional Neural Networks. arXiv: Image and Video Processing.
3. David J. Ma, Yanting Yang, Natalia Harguindeguy, Ye Tian, Jia Guo. Magnetic Resonance Spectroscopy Spectral Registration Using Deep Learning. bioRxiv 2022.08.31.506120; doi: https://doi.org/10.1101/2022.08.31.506120
4. G Oeltzschner, HJ Zöllner, SCN Hui, M Mikkelsen, MG Saleh, S Tapper, RAE Edden. Osprey: Open-Source Processing, Reconstruction & Estimation of Magnetic Resonance Spectroscopy Data. J Neurosci Meth 343:108827 (2020).
5. David J. Ma, Yanting Yang, Natalia Harguindeguy, Ye Tian, Jia Guo. Magnetic Resonance Spectroscopy Spectral Registration Using Deep Learning. bioRxiv 2022.08.31.506120; doi: https://doi.org/10.1101/2022.08.31.506120
Figures