5154
EMPIRE: Elastographic Magnetization Prepared Imaging with Rapid Encoding1Department of Medicine, Massachusetts General Hospital, Boston, MA, United States, 2Department of Biomedical Engineering, University of Delaware, Newark, DE, United States, 3Dartmouth College, Hanover, NH, United States
Synopsis
Keywords: Elastography, Data Acquisition
This study demonstrates the use of magnetization preparation for MR elastography in combination with a 3D GRE stack of spirals pulse sequence. By decoupling the motion encoding from the readouts, rapid encoding strategies, such as 3D GRE, can be used. MRE images, displacement, and inverted elastograms are obtained on in vivo human brain. 3D GRE readouts with EMPIRE motion encoding can acquire a complete MRE dataset 33% faster than 2D SE-EPI with similar parallel imaging acceleration factors.
INTRODUCTION
Magnetic resonance elastography (MRE) measures tissue stiffness noninvasively, enabling comparisons between brain structure, health, and functional status1. This technique is enabled by motion sensitization via the use of phase encoding gradients with mechanical excitation of the tissue of interest. These motion encoding blocks are on the order of the period of applied vibration, nominally multiples of 20-60 ms, followed by imaging readouts to sample k-space and form images. High-resolution MRE imaging strategies often employ multishot k-space sampling2-4 to minimize geometric distortion5. However, multishot approaches that use many readouts, such as in fully 3D sampling, incur a significant temporal cost of playing out a motion encoding block for each readout, making fast 3D acquisitions difficult to implement. Many MRE applications using 3D encoding avoid this by reducing FOV in Z (the slice direction), such as multislab2 or multiband imaging6. Here we propose to decouple motion encoding from the imaging readout to enable more trajectories and rapid encoding to be used with MRE. We demonstrate this by combining motion sensitized magnetization preparation with batched, fast 3D spiral GRE readouts entitled elastographic magnetization prepared imaging with rapid encoding (EMPIRE).METHODS
EMPIRE encompasses a magnetization preparation block consisting of slab selective 90x-180y-90-x RF pulses, with synchronized MRE motion encoding gradients bilateral to the 180 pulse (Figure 1). Once the preparation block is played, all accessible longitudinal magnetization in the imaging volume has the synchronized MRE motion encoded in the phase of the object. Forming an MRE image from this magnetization can use any 2D or 3D sequence without further MRE gradients or synchronization to the mechanical vibration. The object does recover unprepared magnetization (without motion encoding), which would leak into the final image, thus magnitude stabilizers7 were implemented to ensure that only the prepared magnetization was read out in the imaging echo train.The EMPIRE motion encoding strategy was implemented in a full pulse sequence as shown in Figure 1 with a 3D stack-of-spirals gradient echo acquisition scheme. All data was acquired on a 3T Siemens Prisma scanner with a 64-channel head and neck coil. Imaging parameters included: 240x240x120 mm3 FOV, 96 x 96 x48 matrix size, 2.5 mm isotropic resolution, TE/TR for each GRE readout = 1.96/9.6 ms, total TR of 1.323 s, including a pause of 800 ms after readout is complete before the next motion encoding block, spiral readout duration 5 ms, 8 designed in-plane spiral interleaves with 4 sampled for a total reduction factor of 2, with all kz encodes corresponding to each in-plane spiral interleave being acquired in centric order following an encoding block.
Motion was encoded via 70 mT/m MRE gradients at 50 Hz mechanical vibration via an acoustic driver (Resoundant Systems) with a pneumatic head pillow. Vibration was only applied during the shot pause between shots and during the EMPIRE motion encoding block, but not during the imaging readout. 24 total MRE images with 4 phase offsets were acquired.
Acquired data was reconstructed via a conjugate gradient SENSE algorithm with NUFFT penalized weighed least squares algorithm implemented in MATLAB with quadratic regularization. Shear stiffness and damping ratio property maps were obtained from the acquired images via nonlinear inversion (NLI)8.
RESULTS
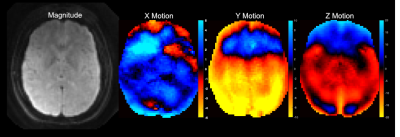
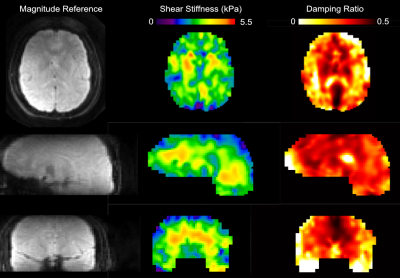
Figure 2 shows the resulting magnitude and phase images from the EMPIRE acquisition. Displacement fields are of high quality and the octahedral-shear strain signal to noise ratio (OSS-SNR)9 was 9.1, which is well above the threshold of 3 required for reliable stiffness parameter inversion via NLI. Figure 3 shows shear stiffness and damping ratio property maps obtained via NLI from the displacements, with strong contrast and anatomical detail.DISCUSSION and CONCLUSIONS
This work demonstrates a proof-of-concept implementation of the novel EMPIRE motion encoding strategy for MRE via 3D GRE readout. Using the sequence we acquired whole-brain MRE data at 2.5 mm isotropic resolution in 2 minutes 10 seconds. For reference, a commonly used 2D SE-EPI MRE sequence with identical coverage and resolution requires 3 minutes 15 seconds to acquire the same data with similar reduction factors.The EMPIRE motion encoding strategy as implemented in this work used 3D stack-of-spiral GRE readouts; however, the choice of readout is independent of the EMPIRE motion encoding. Any suitable 2D or 3D motion encoding strategy could be used, including turbo spin echo (TSE) or 3D spin echo EPI, if the matching magnitude stabilizer rewinder is incorporated into every readout of the sequence. For sequences with many excitations, such as GRE sequences, the loss of prepared magnetization with each excitation results in a loss of peak signal magnitude with each subsequent readout. In the centrically reordered 3D GRE stack of spirals readout used in this work, the loss of signal magnitude per echo results in a Z directed blurring. However, combining a TSE based sequence with magnitude stabilizers7 and the EMPIRE motion encoding strategy is an area of investigation for future work.
Acknowledgements
This project was supported by NIH grants R01-EB027577, R01-AG058853, and U01-NS112120.References
[1] Johnson CL, Schwarb H, Horecka KM, McGarry MDJ, Hillman CH, Kramer AF, Cohen NJ, Barbey AK. Double dissociation of structure-function relationships in memory and fluid intelligence observed with magnetic resonance elastography. Neuroimage. 2018 May 1;171:99-106. doi: 10.1016/j.neuroimage.2018.01.007. Epub 2018 Jan 6. PMID: 29317306; PMCID: PMC5857428.
[2] Johnson CL, Holtrop JL, McGarry MDJ, Weaver JB, Paulsen KD, Georgiadis JG, Sutton BP. 3D Multislab, Multishot Acquisition for Fast, Whole-Brain MR Elastography with High SNR Effiency. Magn Reson Med. 2014 Feb: 71(2): 477-485.
[3] Peng X, Sui Y, Trzasko JD, Glaser KJ, Huston J 3rd, Ehman RL, Pipe JG. Fast 3D MR elastography of the whole brain using spiral staircase: Data acquisition, image reconstruction, and joint deblurring. Magn Reson Med. 2021 Oct;86(4):2011-2024. doi: 10.1002/mrm.28855.
[4] Sui, Y, Arani, A, Trzasko, JD, et al. TURBINE-MRE: A 3D hybrid radial-Cartesian EPI acquisition for MR elastography. Magn Reson Med. 2020; 85: 945– 952. https://doi.org/10.1002/mrm.28445
[5] McIlvain G, McGarry MDJ, Johnson CL. Quantitative effects of off-resonance related distortion on brain mechanical property estimation with magnetic resonance elastography. NMR in Biomedicine. 2022; 35(1):e4616. doi:10.1002/nbm.4616
[6] McIlvain G, Cerjanic AM, Christodoulou AG, McGarry MDJ, Johnson CL. OSCILLATE: A low-rank approach for accelerated magnetic resonance elastography. Magn Reson Med. 2022 Oct;88(4):1659-1672. doi: 10.1002/mrm.29308.
[7] Van AT, Cervantes B, Kooijman H, Karampinos DC. Analysis of phase error effects in multishot diffusion-prepared turbo spin echo imaging. Quant Imaging Med Surg. 2017;7(2):238-250. doi:10.21037/qims.2017.04.01
[8] McGarry MDJ, Van Houten EEW, Johnson CL, Georgiadis JG, Sutton BP, Weaver JB, Paulsen KD. Multiresolution MR Elastography Using Nonlinear Inversion. Medical Physics, 2012; 39(10):6388-6396.
[9] McGarry MD, Van Houten EE, Perriñez PR, Pattison AJ, Weaver JB, Paulsen KD. An octahedral shear strain-based measure of SNR for 3D MR elastography. Phys Med Biol. 2011 Jul 7;56(13):N153-64. doi: 10.1088/0031-9155/56/13/N02.
Figures

Figure 1: (Top) Overview of EMPIRE pulse sequence showing motion encoding and readout blocks with mechanical excitation. (Bottom Left) An illustration of the EMPIRE motion encoding block demonstrating the MRE motion encoding gradients, magnitude stabilizer and slab selective RF pulse with slice select rewinders. (Bottom Right) 3D GRE stack of spirals readout with magnitude stabilizer rewinders prior to spiral readout.