5136
2D U-Net for left heart segmentation for CINE sequences, deal with new target data: comparison of 3 approaches
Habib Rebbah1, Guillaume Gautier1, and Timothé Boutelier1
1Research & Innovation, Olea Medical, La Ciotat, France
1Research & Innovation, Olea Medical, La Ciotat, France
Synopsis
Keywords: Heart, Segmentation
In the case of U-Net based segmentation algorithms, we propose to explore a common question of the medical image research teams: using already existing algorithm or train a new one specific to the target data. We compared for the cardiac CINE case three approaches: using an algorithm trained with external data, train a new one with the taget data from scratch, and fin-tune the first one with the target data.Introduction
While the U-Net1 architecture became the state of the art approach for automatic fast segmentation in medical image processing, global databases with various cases allowing a generalization of the trained algorithms, remain rare. Instead, we often deal with limited database highly specific to the studied pathologies or to the used protocol of a particular medical institution. Here, we propose to explore a common issue for the institutions: using an already available algorithm or design and train a new one specific to their protocols.Methods
We explore the case of left heart segmentation (left ventricle -LV- and myocardium) for CINE sequences in short axis view. Using the MONAI2 framework, we compared three approaches based on U-Net architecture: using an algorithm developed with external data; training a new one with the target data; enhancing the old algorithm with transfer learning. The three approaches are named respectively: ACDC, HIBISCUS, and transfer. Table 1 details the CINE sequences datasets used.The three algorithms were based on the same U-Net architecture and trained with the same ration 90/10% of train/test division of the databases. To compare them, we evaluated them on the HIBISCUS evaluation dataset (38 patients). Figure 1 illustrates the methodology adopted. Finally, to obtain a sharper evaluation, we split the results according to the position of the slice along the long axis. The slice positions were normalized to match the [0 -base-, 100 -apex-] interval.
Results
Figure 2 gathers the results in term of Dice coefficient and Hausdorff distance computed on the HIBISCUS evaluation dataset for the LV and myocardium labels according to the slice position.For all the methods and labels, we observe a lower Dice value (and higher Hausdorff distance) for base and apex slices compared to the mid-cavity ones. We also observe a monotonous decay relation between the slice position and the median Dice values for these ones. However, such a relation is not observed for the Hausdorff distance.
For LV label, except for the apex slices with the ACDC algorithm (0.85), all the median Dice values are higher than 0.93. The ACDC algorithm presents the lowest median Dice values for all the slice positions, while the HIBISCUS and transfer ones are comparable to each other (with less than 0.01 in favor of transfer algorithm).
We observe the same behavior for the myocardium label. The performances are better with the HIBISCUS and transfer methods (with a comparable advantage for transfer one) than with the ACDC one. In term of Dice values: for the mid-cavity slices, the medians of the first algorithms are in ]0.88, 0.93[ interval compared with the ]0.81, 0.87[ interval for ACDC. The worst performances are also observed for the apex slices cases were the median of ACDC is lower than 0.72. However, comparing to the LV label, the transfer algorithm presents the highest performances for these slices, while we observe a higher uncertainty with the HIBISCUS algorithm.
The Hausdorff distance observations bring out the same elements.
Discussion
The advantage of splitting the results according to the slice positions is highlighted in this study. Indeed, the performances were better for the mid-cavity slices where the myocardium is mainly a ring highly delimited by the LV and the surrounded structures. Shuffle all the slices leads to a decrease of the observed performances, whereas those that contribute the most to the marker of interest (here the ejection fraction), present significant better results. The base and apex slices could include a large variety of confusing structures such as atrium walls, large variety of myocardium shape or small apex region highly subject to partial volume effect. The monotonous decay relation between the slice position and the median Dice values for the mid-cavity slices reflects the decrease of the myocardium volume while the basic unit for computing the Dice remain the voxel. Indeed, this relation is not observed for the Hausdorff distance.Despite the differences between the algorithms results, all three present acceptable performance for the slices of interest (median Dice value higher than 0.93). However, the results show that adapting the U-Net to the target data will always increase the quality of the results. They also suggest that the fine tuning of already trained algorithm leads to even better performances than training a new algorithm from scratch with the target data. This is particularly noticeable for the myocardium apex case, where the uncertainty is highly reduced with the transfer algorithm.
Conclusion
The medical image processing research teams often face a similar question: for the purpose of a specific issue, do we need to adapt old algorithms to our new data. In the case of a U-Net for segmentation, the used data to train the old algorithm are often inaccessible.Depending on the access to the data, ability to fine tuning an existing algorithm, and available time, it could be more or less advantageous to choose one of the three proposed approaches. However, if the conditions are met, this study suggests that the best approach is to fine-tune an existing algorithm with the target data.
Acknowledgements
This work was supported by the RHU MARVELOUS (ANR-16-RHUS-0009). The authors particularly thank Pierre Croisille, Magalie Viallon, Nathan Mewton, Charles De Bourguignon, and Lorena Petrusca for data acquisition and management.References
1. Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 (eds. Navab, N., Hornegger, J., Wells, W. M. & Frangi, A. F.) 234–241 (Springer International Publishing, 2015). doi:10.1007/978-3-319-24574-4_28.
2. MONAI. https://monai.io/about.html.
Figures

Table 1 - Cardiac CINE
sequences databases. ACDC: used for training the existing algorithm; HIBISCUS: target
data.
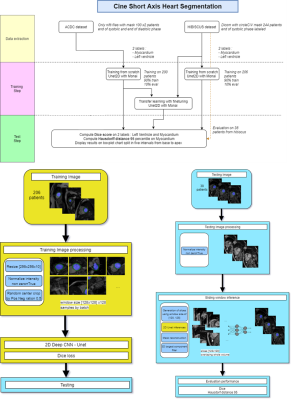
Figure 1 - Cardiac
CINE in short axis: U-Net based segmentation.
Methodology for the 3 approaches.
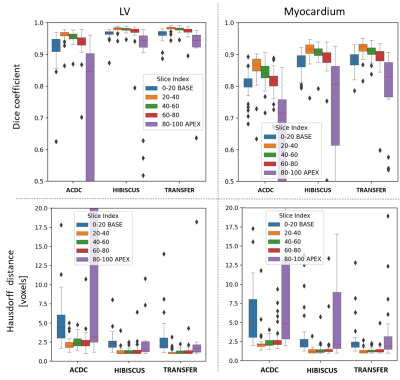
Figure 2 - Cardiac CINE segmentation: results. Comparing
the three approaches on the evaluation dataset of HIBISCUS (38 patients). Up: Dice coefficients. Down: Hausdorff distances. Left: LV label. Right: myocardium label.
DOI: https://doi.org/10.58530/2023/5136