5132
Deep Learning-Based Reconstruction of Accelerated Cardiac Cine MRI at 0.55T1Computational Imaging Lab, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 2Magnetic Resonance, Siemens Healthcare GmbH, Erlangen, Germany, 3Institute of Radiology, University Hospital Erlangen, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 4Siemens Medical Solutions USA Inc., Chicago, IL, United States, 5Biomedical Engineering, The Ohio State University, Columbus, OH, United States
Synopsis
Keywords: Heart, Low-Field MRI
Acceleration of cardiac cine MRI is highly desirable in order to decrease the required breath-hold duration. On low-field MRI systems in particular, this could help make cardiac MRI more widely available. We present a method based on the Variational Network for reconstruction of cardiac cine MRI, trained on data from 1.5 and 3T systems. Reconstructions of retrospectively and prospectively undersampled acquisitions at 0.55T with an acceleration rate of eight are shown and compared to Compressed Sensing reconstructions. Despite the domain shift to low-field data, the neural network achieved an SSIM of 94.6%, which is comparable to the Compressed Sensing results.
Introduction
Cardiac cine MRI commonly requires long breath-holds that may not be feasible for all subjects. Scan times at low magnetic field strengths can additionally be prolonged due to system capabilities and inherently lower signal-to-noise ratio. Considering the advantages of lower field strengths in cardiac MRI1, accelerated acquisition is highly desirable to make CMR widely available. While classical Parallel Imaging techniques like GRAPPA and SENSE are limited in acceleration and hence in temporal resolution, Compressed Sensing (CS) and, more recently, Deep Learning may be suitable reconstruction techniques in this setting. For Deep Learning, however, either enough corresponding training data is required, or a domain shift must be bridged by the network if trained with different data.We present Deep Learning-based reconstructions of accelerated cardiac cine acquisitions on a 0.55T MRI system and compare them to CS with total variation (TV) regularization.
Methods
Fully sampled cardiac cines of seven healthy volunteers were acquired with a research sequence on a 0.55T scanner (MAGNETOM Free.Max, Siemens Healthcare, Erlangen, Germany) with a 15 coil element array. Acquisition matrix sizes were between 192x133 and 192x180 (field of view between 360mm x 270mm and 360mm x 345mm), number of frames between 16 and 28 and temporal resolution ranged from 30.5ms to 51.4ms. Of the 41 slices acquired, 30 were short-axis views, six two-chamber, and five four-chamber views. Per slice, a 16-22s breath-hold was required. This data was retrospectively undersampled by a factor of eight using an incoherent variable density sampling mask.Prospectively undersampled data was acquired from three volunteers using the same incoherent sampling pattern. In total, 20 cines (14 short-axis, three two-chamber, and three four-chamber views) with 26-35 frames and temporal resolution between 29.1ms and 36.2ms were acquired over the duration of three heartbeats each.
CS reconstruction was performed using Bart2 with TV regularization over time3. Regularization parameters were chosen such that both aliasing artifacts and impact on motion fidelity were minimized. Coil sensitivities were estimated from time-averaged k-space using ESPIRiT4.
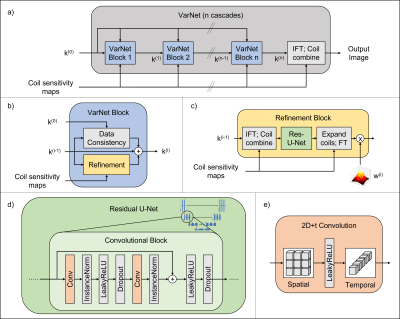
Deep learning-based reconstruction was performed with a spatiotemporal neural network based on the End-to-End Variational Network5,6 with 15 cascades (cf. Figure 1). It differs from the original architecture in that it contains residual instead of plain U-Nets and uses spatiotemporal convolutions to exploit correlations over time. Additionally, the regularization term was scaled with a learnable piecewise linear function to allow the network to focus on certain areas of k-space in each cascade. Furthermore, the same precomputed coil sensitivity profiles were used as for the CS reconstructions.
The network was trained using fully sampled cine raw data from 1.5 and 3T scanners from the OCMR dataset7. Apart from the field strength, this data differs from the acquired low-field data in the number of coils (15-38) and matrix size (144x112-256x208). The number of frames and temporal resolutions are in a similar range as in the fully sampled low-field data. 142 cine slices from 33 acquisitions were used for training and 22 single-slice acquisitions for validation. Retrospectively undersampled k-space data was used as input and sensitivity-weighted coil-combined images of the fully sampled data as ground truth. A combination of SSIM loss and a loss function penalizing the reconstruction’s phase component, the so-called ⊥-loss8, was used. The network was trained for 100 epochs using an Adam optimizer9. The trained network was then applied on both the retrospectively and the prospectively undersampled low-field data.
Results
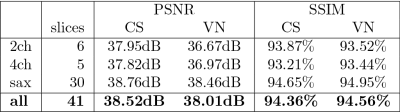
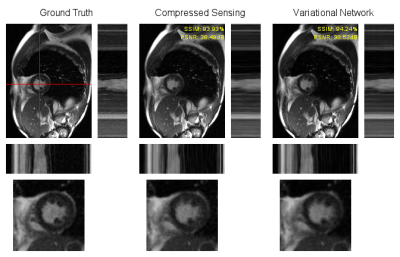
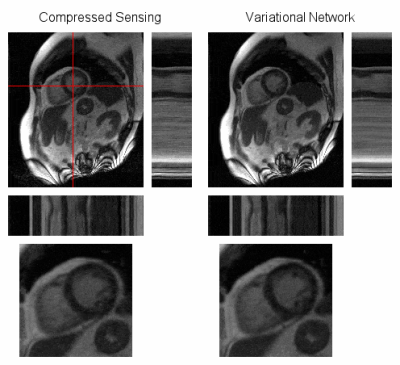
Training took ~5h on three A100 GPUs in parallel and reconstruction time during inference was 1.5-2s per cine.On average, the network achieved a structural similarity (SSIM) of 94.56% and peak signal-to-noise ratio (PSNR) of 38.01dB on the retrospectively undersampled low-field cines. With CS, an average SSIM of 94.36% and PSNR of 38.52dB were achieved. Detailed quantitative results are summarized in Figure 2. Visually, both methods achieved good reconstruction results with only slight residual artifacts. Exemplary reconstructions are illustrated in Figures 3-4. Reconstructions of short-axis slices were generally better than those of two- and four-chamber views visually and in terms of image metrics.
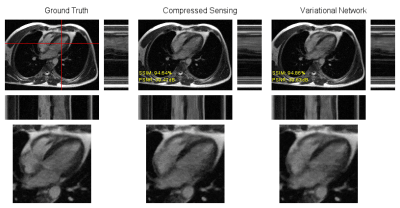
The reconstructions of the prospectively undersampled data with both the neural network and CS contain noticeable residual aliasing artifacts, but cardiac motion was well restored. Figure 5 shows exemplary reconstructions.
Discussion
The spatiotemporal Variational Network showed good reconstruction performance on both retrospectively and prospectively undersampled cardiac cines at 0.55T with reconstruction times that are appropriate for clinical practice. The network was able to bridge the domain shift from 1.5 and 3T to 0.55T, which could be even further improved if enough fully sampled low-field data is available to train the network using this data. However, this is challenging due to long breath-holds required for acquisition.The performance difference between short-axis and long-axis views might be attributed to an imbalance in the training data, containing 128 short-axis but only 14 long-axis slices. Furthermore, the slight performance drop from retrospectively to prospectively undersampled acquisitions highlights the necessity for prospective studies.
Conclusion
We have demonstrated Deep Learning-based reconstructions of both retrospectively and prospectively accelerated cardiac cines at low field with similar reconstruction quality as CS. Considering the breath-hold duration of three heartbeats and properties of low-field MRI systems, the presented method could help make cardiac MRI more widely accessible.Acknowledgements
This research was supported by NIH/NIBIB grant R01EB029957.
References
1. Campbell-Washburn AE, Ramasawmy R, Restivo MC, et al. Opportunities in Interventional and Diagnostic Imaging by Using High-Performance Low-Field-Strength MRI. Radiology 2019;293:384–393.
2. Uecker M, Rosenzweig S, Holme M, et al. mrirecon/bart: version 0.6.00; 2020.
3. Feng L, Axel L, Chandarana H, et al. XD‐GRASP: Golden‐angle radial MRI with reconstruction of extra motion‐state dimensions using compressed sensing. Magn Reson Med 2016;75:775–788.
4. Uecker M, Lai P, Murphy MJ, et al. ESPIRiT-an eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA. Magn Reson Med 2014;71:990–1001.
5. Vornehm M, Wetzl J, Giese M, et al. Spatiotemporal variational neural network for reconstruction of highly accelerated cardiac cine MRI. Eur Heart J - Card Img. 2022;23(Supplement_2):jeac141.018
6. Sriram A, Zbontar J, Murrell T, et al. End-to-End Variational Networks for Accelerated MRI Reconstruction. In: Martel AL, et al., editors. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. Cham: Springer International Publishing; 2020. pp. 64–73.
7. Chen C, Liu Y, Schniter P, et al. OCMR (v1.0)--Open-Access Multi-Coil k-Space Dataset for Cardiovascular Magnetic Resonance Imaging. arXiv:2008.03410 [eess] 2020.
8. Terpstra ML, Maspero M, Sbrizzi A, et al. ⊥-loss: A symmetric loss function for magnetic resonance imaging reconstruction and image registration with deep learning. Medical Image Analysis 2022;80:102509.
9. Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. arXiv:1412.6980 [cs] 2017.
Figures