5130
Detection of Congenital Heart Disease in MR images using Machine Learning1Department of Congenital Heart Disease and Pediatric Cardiology, University Hospital Schleswig-Holstein, DZHK (German Center for Cardiovascular Research), partner site Hamburg/Kiel/Lübeck, Kiel, Germany, 2Multimedia Information Processing Group, Kiel University, Kiel, Germany, 3Departments of CMR and Paediatric Cardiology, Royal Brompton Hospital, London, United Kingdom
Synopsis
Keywords: Heart, Machine Learning/Artificial Intelligence, Congenital Heart Disease
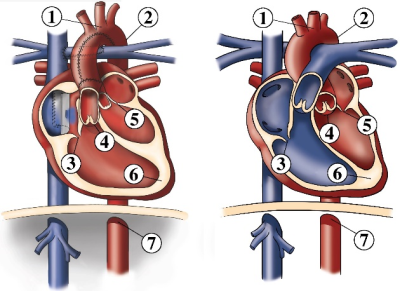
We present a new method for detection of hypoplastic left heart syndrome (HLHS) based on the spatial arrangement of 7 distinctive anatomical landmarks in CMR images. The method was applied to the axial SSFP CMR scans of 46 patients with HLHS and 33 healthy controls. A tailor-made U-net-like deep convolutional network (CNN) with a shared 3D-convolutional encoder backbone and 7 segmentation heads was used for prediction of landmarks. Classification based exclusively on the coordinates of the detected landmarks had an accuracy of 98.7%. In future studies, the method may be applied to HLHS subgroups or other cardiac diseases.Purpose
In recent years, the great potential of artificial intelligence to support clinical evaluation of cardiovascular MRI (CMR) data has become increasingly apparent [1, 2]. Several CMR studies applied machine learning to congenital heart disease to address specific conditions related to segmentation [3], assessment of surgical results [4], prediction of prognosis [5, 6], hierarchical clustering of congenital heart disease [7] and data augmentation [8, 3]. Machine learning classification of complex congenital heart disease has been demonstrated using echocardiography [9] but only few studies exist that have used CMR for classification of complex congenital heart disease using machine learning [7].Methods
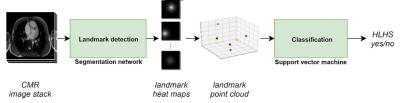
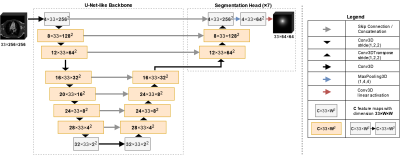
This study presents a new method for detection of the hypoplastic left heart syndrome (HLHS) based on the spatial arrangement of 7 distinctive anatomical landmarks in CMR images (Figure 1). The method was applied to the axial SSFP CMR scans of 46 patients with HLHS and 33 healthy controls. A flow chart of the method is presented in Figure 2. A tailor-made U-net-like deep convolutional network (CNN) with a shared 3D-convolutional encoder backbone and 7 segmentation heads was used for prediction of the landmarks (Figure 3). For training and evaluation purposes, manual annotation of the landmarks was prepared by two observers. Cross-validation was performed with 5 folds, which split data into training and validation data in a ratio 80:20. Standard deviation between prediction and annotation was determined for each landmark and compared to the interobserver variability. In the final step, the actual classification was based exclusively on the coordinates of the detected landmarks and was performed by a linear support vector machine (SVM). The classification accuracy was evaluated for a variable depth (between 2 and 6) of 7 segmentation heads and the use of augmentation for training of the CNN and the SVM.Results
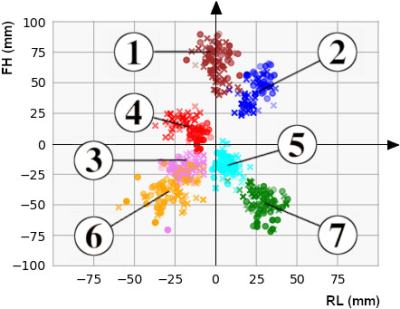
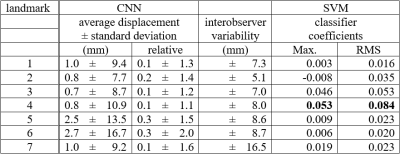
Distributions of annotated landmarks after centering are depicted in figure 4. Training of one segmentation network took 20 minutes. The final neural network architecture had 6 filters in the first layer and applied a splitting into segmentation heads above depth 3. No augmentation was used for training. Details about the hyperparameter search are described at the end of the results section. Average displacement and standard deviation between predicted and annotated landmarks as well as the interobserver variability are shown in Table 5. The largest displacements and standard deviations were reached for landmarks 6 (apex, displacement 2.7 mm, standard deviation 16.7 mm) and 5 (mitral valve, displacement 2.5 mm, standard deviation 13.5 mm). For other landmarks, average displacements were typically in the order of 1 mm and standard deviations were below 11 mm. Also, in comparison with the interobserver variability, the detection of the right ventricular apex (landmark 6) had the highest deviations (2.0 times higher than the interobserver variability). Deviations of all other landmarks had about the size of the interobserver variability (ratios ranging between 1.1 and 1.6). Training of the classifier on manual annotation training data took less than 1 minute. When skipping the landmark detection and using manual annotation test data directly as input to classifier prediction, the classification accuracy was 100.0%. All datasets were correctly classified. After landmark detection using the final hyperparameters, the average classification accuracy was 98.7%. All but one dataset were correctly classified. Coefficients learned by the SVM are listed in Table 5. The coefficients for the aortic valve landmark had the largest values followed by the tricuspid valve. Changing the number of filters in the first layer from 6 to 3 (9) changed the classification accuracy to 96.2% (89.9%). The variation of the depth of the segmentation heads from 4 down to 2 or up to 6 had no effect on the classification accuracy of 98.7%. Using data augmentation for training the neural network had no further effect on the classification accuracy. Also, augmentation of data used for classification had no further effect on the accuracy.Conclusion
This study presents a new method for machine-learning-based identification of HLHS patients after TCPC completion in CMR datasets based on the geometric arrangement of anatomical landmarks. The classification accuracy of the full method was 98.7%, whereas the final step of classification had an accuracy of 100.0% when using manually annotated landmarks. The method was limited by the uncertainty of landmark detection using a tailor-made deep neural network. The prospects of being able to automatically gain information relevant for CMR image analyses opens up new potentials to assist the evaluating physician [9, 10]. In particular, the classification of the cardiovascular anatomy can be an important contribution to make analysis programs smarter. Before examining the images, the physician may be informed about special anatomical conditions. Furthermore, the information can be used to guide the physician more effectively through the analysis which may ultimately improve the work flow and may lead to a time saving regarding CMR image assessment. In future studies, the method may be applied to HLHS subgroups or other cardiac diseases.Acknowledgements
No acknowledgement found.References
1. Fotaki A, Puyol-Antón E, Chiribiri A, Botnar R, Pushparajah K, Prieto C. Artificial Intelligence in Cardiac MRI: Is Clinical Adoption Forthcoming? Front Cardiovasc Med. 2022; 8:818765.
2. Helman SM, Herrup EA, Christopher AB, Al-Zaiti SS. The role of machine learning applications in diagnosing and assessing critical and non-critical CHD: a scoping review. Cardiol Young. 2021; 31(11):1770-1780.
3. Karimi-Bidhendi S, Arafati A, Cheng AL, Wu Y, Kheradvar A, Jafarkhani H. Fully‑automated deep‑learning segmentation of pediatric cardiovascular magnetic resonance of patients with complex congenital heart diseases. J Cardiovasc Magn Reson. 2020; 22(1):80.
4. Lu Y, Fu X, Li X, Qi Y. Cardiac Chamber Segmentation Using Deep Learning on Magnetic Resonance Images from Patients Before and After Atrial Septal Occlusion Surgery. Annu Int Conf IEEE Eng Med Biol Soc. 2020; 2020:1211-1216.
5. Diller GP, Orwat S, Vahle J, Bauer UMM, Urban A, Sarikouch S, Berger F, Beerbaum P, Baumgartner H; German Competence Network for Congenital Heart Defects Investigators. Prediction of prognosis in patients with tetralogy of Fallot based on deep learning imaging analysis. Heart. 2020; 106(13):1007-1014.
6. Diller GP, Kempny A, Babu-Narayan SV, Henrichs M, Brida M, Uebing A, Lammers AE, Baumgartner H, Li W, Wort SJ, Dimopoulos K, Gatzoulis MA. Machine learning algorithms estimating prognosis and guiding therapy in adult congenital heart disease: data from a single tertiary centre including 10 019 patients. Eur Heart J. 2019; 40(13):1069-1077.
7. Bruse JL, Zuluaga MA, Khushnood A, McLeod K, Ntsinjana HN, Hsia TY, Sermesant M, Pennec X, Taylor AM, Schievano S. Detecting Clinically Meaningful Shape Clusters in Medical Image Data: Metrics Analysis for Hierarchical Clustering Applied to Healthy and Pathological Aortic Arches. IEEE Trans Biomed Eng. 2017; 64(10):2373-2383.
8. Diller GP, Vahle J, Radke R, Vidal MLB, Fischer AJ, Bauer UMM, Sarikouch S, Berger F, Beerbaum P, Baumgartner H, Orwat S; German Competence Network for Congenital Heart Defects Investigators. Utility of deep learning networks for the generation of artificial cardiac magnetic resonance images in congenital heart disease. BMC Med Imaging. 2020; 20(1):113.
9. Chauhan D, Anyanwu E, Goes J, Besser SA, Anand S, Madduri R, Getty N, Kelle S, Kawaji K, Mor-Avi V, Patel AR. Comparison of machine learning and deep learning for view identification from cardiac magnetic resonance images. Clin Imaging. 2022; 82:121-126.
10. Ho N, Kim YC. Evaluation of transfer learning in deep convolutional neural network models for cardiac short axis slice classification. Sci Rep. 2021; 11(1):1839.
Figures