5118
Density-Controllable Spiral Trajectory Design Combined with CS-UTE Improves High-Resolution Single Breath-Hold Lung Imaging
Yupeng Cao1, Jun Zhao1, Weinan Tang2, Wentao Liu1, and Dong Han1
1National Center for Nanoscience and Technology, Beijing, China, 2Wandong Medical Inc, Beijing, China, Beijing, China
1National Center for Nanoscience and Technology, Beijing, China, 2Wandong Medical Inc, Beijing, China, Beijing, China
Synopsis
Keywords: New Trajectories & Spatial Encoding Methods, Lung
Single breath-hold high-resolution lung imaging is challenging due to the short T2* and scan efficiency. The stack-of-spirals UTE enables fast lung imaging in a single breath hold. However, fast scanning of stack-of-spirals results in a long readout time, introducing adverse effects of the short T2* of the lung. Herein, we proposed a density-controllable spiral trajectory (DCST) design method to design a short readout time trajectory concurrently satisfying the criteria of optimal compressed sensing (CS), reducing the adverse effect of the short T2*. The short readout time trajectory combined with CS-UTE improves the single breath-hold high-resolution lung imaging.Introduction
Stack-of-spirals UTE using the less shot numbers allows lung imaging under single breath-hold acquisition (1-2). However, the readout time of the stack-of-spirals is long under high-resolution single breath-hold imaging, which gives rise to adverse effects resulting from the short T2* relaxation time of the lung, including blurring and signal-to-noise ratio decreasing (3). In this study, we propose a density-controllable spiral trajectory design method to reduce the readout time in stack-of-spirals UTE. This method includes two parts: (i) the high-sampling-rate numerical multi-density spliced path, which satisfies optimal CS criteria and (ii) the gradient-constrained discrete k-space trajectory pixel-wise selection. The reconstruction was implemented by CS (4). The UTE sequence employs a short SLR pulse and slice-selective gradient for signal excitation of the slab region, followed by a gradient merged by the slice-rephasing and slice encoding gradient, and finally uses the spiral readout gradient. In addition, the phantom experiment and one healthy volunteer lung imaging were implemented.Methods
This study was approved by the National Center for Nanoscience and Technology Ethics Board, and written informed consent was given by all study participants. All MR images were obtained with a 1.5-T scanner (Wandong Medical Inc, Beijing, China). The single-channel body coil was used in the phantom imaging. The six-channel chest coil was used in the volunteer lung imaging. The DCST design method includes two parts shown in Figure 1 (a). First, towards the targeted density, the analytic formula of the spiral (Equation [1] and [2]) was used to generate a high-sampling-rate numerical path by regulating the deviation of r and the deviation of φ. Each density region was smooth-like spliced. Second, according to the allowed gradient amplitude and slew rate, the k-space trajectory was searched pixel by pixel along the high-sampling-rate numerical path.x=rcos(ϕ) [1]
y=rsin(ϕ) [2]
The undersampling trajectory (32 arms), the long readout time trajectory (32 arms), and the short readout time trajectory (64 arms) were designed as shown in Figure 1 (b). The full sampling was performed in the core region of the k-space, and the multi-density undersampling was performed in the peripheral area of the k-space. The maximum gradient amplitude and slew rate used in this research were 27 mT/m and 60 T/m/s, respectively. The UTE sequence is based on a stack-of-spirals frame. As shown in Figure 2, a 100 μs SLR pulse was employed, followed by a gradient which was merged by the slice rephasing gradient and the slice encoding gradient. First, the 30cm slice thickness region was excited. Then, the spiral gradient was implemented, and the signal was acquired concurrently. The CS reconstructed method used in our experiments has been published by Lustig et al. (4). The large ACR phantom was used in this experiment, as shown in Figure 3. The lung imaging of the healthy volunteer is shown in Figure 4. The volunteer was required to breath-hold in each scan. The scan parameters are shown in Table 1.
Results
Images of the ACR phantom in Figure 3 show that the short-readout time full sampling performed best. The long-readout time full sampling performed worst. The direct Fourier reconstructed undersampling image shows blurring but preserves the structure. The CS-reconstructed undersampling image improves the blurring and appears closer to the image of the short-readout time full sampling. In Figure 4, the scan time of the undersampling lung imaging is the shortest. Clear distinction in lung structure is shown in the CS-reconstructed image, while the lung image of the long readout time full sampling performs ambiguous structure. The short readout time full sampling appeared middle in lung imaging.Discussion
Herein, we presented a spiral trajectory design method, which can control the density in radial and angular directions of k-space. The density-controllable pattern is suitable for the optimal CS criteria. Regarding the contradiction between the readout time and the scan time of stack-of-spirals UTE, this method provided a significantly reduced readout time trajectory and further improved the blurring in single breath hold lung imaging for high resolution. The lung imaging of the undersampling CS reconstruction performed slightly better than the short readout time full sampling, maybe because the scan time of 38.4 seconds is hard for a single breath hold. Although, the slice encoding efficiency is low in our experiments, which leads to the long TE. Our method can be implemented in other high-efficiency slice encoding patterns, including acquisition-weighted UTE (5). Noteworthy, the design method can be extended to different approximately continuous trajectory designs.Conclusion
The proposed method provides an easy approach to designing a trajectory in which the density is controllable in both radial and angular directions. In addition, shortening readout time and CS reconstruction improve the blurring and signal-to-noise ratio of high-resolution lung imaging in single breath-hold acquisition.Acknowledgements
This work was supported by National Natural Science Foundation of China (NO.61971151) and Wandong Medical.References
(1) Mugler JP et al. Proc Intl Soc Mag Reson Med 2015; 23:1476.
(2) Zeimpekis K et al. Proc Intl Soc Mag Reson Med 2016; 24:1608.
(3) Willmering MM, Robison RK, Wang H, Pipe JG, Woods JC. Implementation of the FLORET UTE sequence for lung imaging. Magnetic Resonance in Medicine 2019; 82:1091–1100.
(4) Lustig M, Donoho D, Pauly JM. Sparse MRI: The Application of Compressed Sensing for Rapid MR Imaging. Magnetic Resonance in Medicine 2007; 58:1182–1195.
(5) Qian YX, Boada FE. Acquisition-weighted stack of spirals for fast high-resolution three-dimensional ultra-short echo time MR imaging. Magnetic Resonance in Medicine 2008; 60:135–145.
Figures
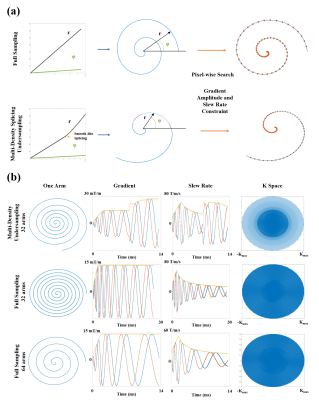
Figure 1: Scheme of the DCST design method and
trajectories of the experiments. (a) The high-density numerical path was
generated by regulating the radial and angular velocity. According to the
allowed gradient amplitude and the slew rate, the optimal k space trajectory
was searched pixel by pixel along the path. (b) Three trajectories were designed:
the undersampling (multi-density in radial and angular directions), the
long-readout time, and the short-readout time full sampling.
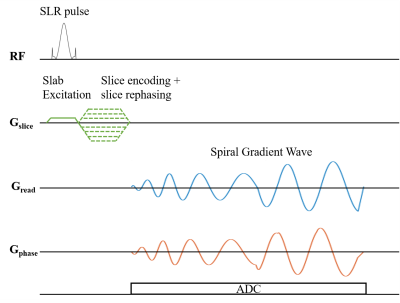
Figure 2: Stack-of-spirals UTE Sequence.
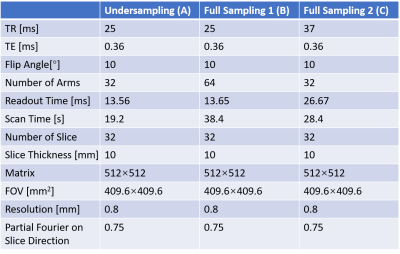
Table 1: Scan parameters for all acquisitions.
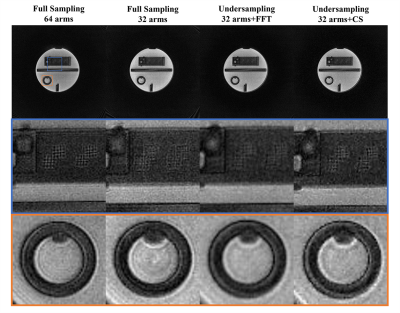
Figure 3: ACR phantom imaging. FFT is fast Fourier
transform reconstruction. The CS is compressed sensing reconstruction.
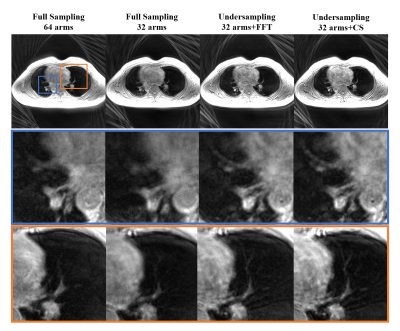
Figure 4: Human lung imaging. The
CS-reconstructed image shows the clearest detail of the lung.
DOI: https://doi.org/10.58530/2023/5118