5110
Multi-contrast multi-resolution UTE for simultaneous quantitative and high-resolution MRI1Division of Medical Physics, Department of Radiology, University Medical Center Freiburg, Freiburg, Germany, 2Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN, United States, 3School of Health Science Department, Purdue University, West Lafayette, IN, United States
Synopsis
Keywords: New Trajectories & Spatial Encoding Methods, New Trajectories & Spatial Encoding Methods
A multi-contrast multi-resolution UTE sequence was developed with a dual-echo acquisition between 1-1 binomial water excitation pulses and a conventional UTE readout. This acquisition scheme provides 3 images with different echoes, flip angles, spatial resolution, and contrast (water excitation). T2* was quantified in the whole brain in TA<15min, and UTE images of the brain were reconstructed with 0.5mm isotropic resolution without additional scan time.
Introduction
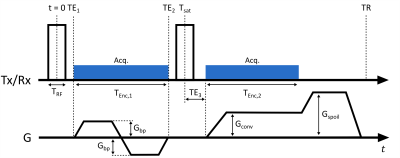
Fat suppression techniques are used in UTE imaging to reduce chemical shift artefacts and to improve dynamic range of water content1. Visualization of short-T2* components can be improved significantly by long-T2 or fat suppression2. Fat saturation with spectrally selective saturation pulses, inversion recovery techniques, and water-selective excitation using binomial pulses have been successfully integrated to UTE sequences3,4.Using composite binomial RF pulses and suitable inter-pulse delays leads to efficient water and fat selective imaging6. The shortest binominal pulse, a 1-1 binomial hard pulse pair, was used for rapid water-excitation in UTE7. At 3T the frequency offset between fat and water is 370-430Hz, which requires an inter-pulse delay of 1.15-1.35ms between the 1-1 binomial pulses. In this study, we integrate an additional lower-resolution dual-echo acquisition between the binomial pulses to make effective use of the 1-1 inter-pulse delay(Fig.1). This acquisition scheme results in 2+1 images with different echoes, flip angles, and spatial resolution, and one of them with water excitation. We investigate using the proposed multi-contrast UTE(mcUTE) sequence in mapping of short-T2* components of brain. The dual echoes between the 1-1 pulses can provide data with different T2* weighting, and the high-resolution fat-suppressed UTE image from echo-3 provides an anatomical reference and might improve the reconstruction.
Methods
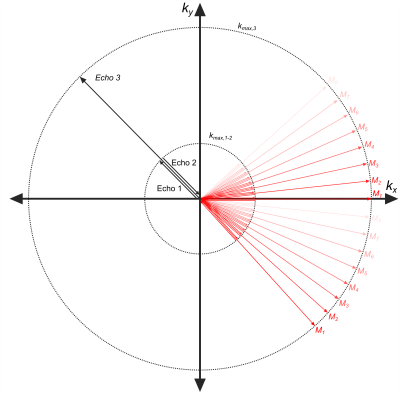
The 3D mcUTE sequence was implemented on a clinical 3T MRI system (Siemens PRISMA-Fit). For 1-1 binomial excitation, 20μs-long hard pulses were used separated by a delay of 1.23 ms (Tsat = 1.25 ms)(Fig. 1). Between the 1-1 pulse, a bipolar gradient pair was inserted with a balanced 0th moment to achieve an effective water excitation. Two echoes were acquired in the presence of the bipolar gradients with the radial acquisitions starting(echo-1) and ending(echo-2) at the k-space center. An encoding time of 450µs was used for each echo. Even though ramp-sampling was applied, the spatial resolution achievable with these two echoes was significantly lower than that of the subsequent UTE acquisition(echo-3), which was only limited by TR and peripheral nerve stimulation(PNS).mcUTE was tested in 2 healthy volunteers using the standard 20-channel head coil of the system for data acquisition. An MP-RAGE image was acquired for reference. The parameters for the mcUTE acquisition for the first/second/third echoes were: FOV=(192mm)3, 1.7mm/1.7mm/0.5mm isotropic voxel-size, FA=4°/4°/8°, TR=2.8ms, Gconv=36mT/m, Gbp=20mT/m, Tenc,2=740us, TRF=20µs, 40000 spokes. To be able to calculate T2* values of the ultra-short T2* components, the measurement was repeated 8 times(M1-M8) with TE1={40,60,80,100,140,180,220,260}µs, TE2=TE1+900µs, and TE3=20µs resulting in a total scan time of 15min. To achieve an (0.5mm)³ isotropic resolution in the 3rd echo with higher SNR, in total 320000 spokes were acquired in a time-interleaved order, where each 40000 spokes acquired during each mcUTE acquisition(Mi,i=1,…,8) were merged for reconstruction retrospectively(Fig.2).
Radial raw data were re-gridded onto a Cartesian grid after density compensation, a Hamming filter was applied to reduce ringing artifacts, and a 3D fast Fourier transform was used to calculate the 3D image data. T2* maps were calculated by pixel-wise fitting the signals from the different echoes to a double -exponential decay curve: $$$S(\text{TE})=S_0e^{\text{TE}/T_{2,short}^*}+S_1e^{\text{TE}/T_{2,long}^*}$$$.
Results
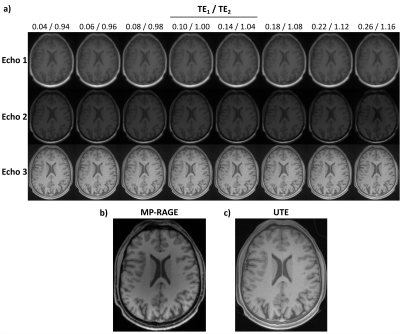
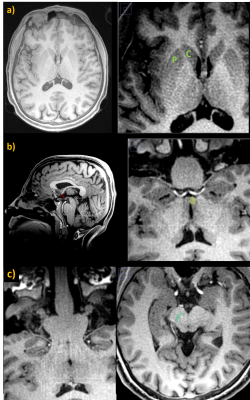
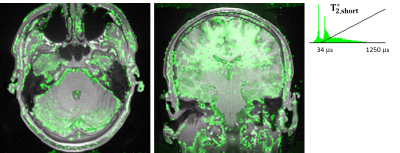
In Fig.3a, UTE images of the first volunteer for varying TEs are shown. The last row in Fig3a shows images from the 3rd echo before merging their corresponding k-spaces. In Fig.3c, the 0.5mm resolution water-excited image reconstructed by retrospectively merging the interleaved k-space data of measurements from the 3rd echoes is shown. In the high-resolution UTE image, deep brain nuclei are easily identifiable such as putamen and caudate, which together form the striatum. Water excitation by the 1-1 pulse provided additional contrast in these nuclei by suppressing the lipid content (Fig.4). Another example is the mammillary body, which is clearly visible in the high-resolution data without any distortions (Fig.4b).In Fig.5, the T2* maps showing short T2 components in the brain of each volunteer are presented. Short T2* component values were intersubject consistent. For example T2*(pons)=95.6±8.0/112.7±15.9μs; T2*(putamen)=174.7±27.5/133.7±9.4μs; T2*(red nucleus)=143.3±23.7/126.7±25.0μs; T2*(anterior-lobe-of-cerebellum)=101.8±12.8/97.5±11.4μs for Volunteer 1/2, respectively.
Discussion
In this work, we integrated an additional dual-echo UTE encoding between the binomial pulses to take advantage of the time between 1-1 pulses and calculated a map of short-T2* components of brain. As the sequence uses different flip angles and encoding schemes, it could also be used for other applications. For example, bipolar gradients could also be applied in the UTE section so that 4 echoes are acquired. While echo-1 and 2 would have one flip angle $$$\alpha$$$, the UTE echoes 3 and 4 would be acquired with $$$2\alpha$$$ from the water excitation. Such a framework can also be used for simultaneous T2* and T1 mapping using the FLASH equation.Information provided by the high-resolution fat-suppressed UTE data can be used for interpreting the quantitative data, improving the reconstruction of the dual-echo images by, e.g., joint reconstruction techniques8, for better segmentation of skull, or for better segmentation of deep brain nuclei, as they are mostly separated by lipid-containing membranes.
Conclusion
In this work, we presented a new UTE sequence with multi-contrast and multi-resolution encoding. Using a total scan time of 15 minutes, T2* components of the whole brain volume were quantified. In addition to the T2* maps, the sequence provided a very high-resolution UTE image of the brain.Acknowledgements
.References
(1) Ma, Y.; Jang, H.; Jerban, S.; Chang, E. Y.; Chung, C. B.; Bydder, G. M.; Du, J. Making the Invisible Visible—Ultrashort Echo Time Magnetic Resonance Imaging: Technical Developments and Applications. Appl. Phys. Rev. 2022, 9 (041303), 1–40. https://doi.org/10.1063/5.0086459.
(2) Gatehouse, P. D.; Bydder, G. M. Magnetic Resonance Imaging of Short T2 Components in Tissue. Clin. Radiol. 2003, 58 (1), 1–19. https://doi.org/10.1053/crad.2003.1157.
(3) Afsahi, A. M.; Ma, Y.; Jang, H.; Jerban, S.; Chung, C. B.; Chang, E. Y.; Du, J. Ultrashort Echo Time Magnetic Resonance Imaging Techniques: Met and Unmet Needs in Musculoskeletal Imaging. J. Magn. Reson. Imaging 2022, 55 (6), 1597–1612. https://doi.org/10.1002/jmri.28032.
(4) Du, J.; Bydder, M.; Takahashi, A. M.; Carl, M.; Chung, C. B.; Bydder, G. M. Short T2 Contrast with Three-Dimensional Ultrashort Echo Time Imaging. Magn. Reson. Imaging 2011, 29 (4), 470–482. https://doi.org/10.1016/j.mri.2010.11.003.
(5) Bley, T. A.; Wieben, O.; François, C. J.; Brittain, J. H.; Reeder, S. B. Fat and Water Magnetic Resonance Imaging. J. Magn. Reson. Imaging 2009, 31 (1), 4–18. https://doi.org/10.1002/jmri.21895.
(6) Schick, F. Simultaneous Highly Selective MR Water and Fat Imaging Using a Simple New Type of Spectral-Spatial Excitation. Magn. Reson. Med. 1998, 40 (2), 194–202. https://doi.org/10.1002/mrm.1910400205.
(7) Springer, F.; Steidle, G.; Martirosian, P.; Grosse, U.; Syha, R.; Schabel, C.; Claussen, C. D.; Schick, F. Quick Water-Selective Excitation of Fast Relaxing Tissues with 3D UTE Sequences. Magn. Reson. Med. 2014, 71 (2), 534–543. https://doi.org/10.1002/mrm.24684.
(8) Kopanoglu, E.; Güngör, A.; Kilic, T.; Saritas, E. U.; Oguz, K. K.; Çukur, T.; Güven, H. E. Simultaneous Use of Individual and Joint Regularization Terms in Compressive Sensing: Joint Reconstruction of Multi-Channel Multi-Contrast MRI Acquisitions. NMR Biomed. 2020, 33 (4), e4247. https://doi.org/10.1002/nbm.4247.
Figures