5103
Synthesizing CT Image from Single-echo UTE-MRI using Multi-Task Framework1Biomedical Engineering, Columbia University, New York City, NY, United States, 2Columbia University, New York City, NY, United States, 3Neuroscience, Columbia University, New York City, NY, United States
Synopsis
Keywords: Image Reconstruction, Multimodal
This abstract proposed a cross-modality conversion method from UTE MRI to CT images. Using the TABS and ResidualAttentionU-net model in a processing framework combining image segmentation and prediction, the skull structure can be extracted from UTE MRI with high similarity of CT based skull. Five UTE-CT image pairs of mouse brains were used in the study. And a 3D-patch based training strategy was adopted, which took the advantage of structural continuity between slices in very limited datasets. The results show that the proposed combined image segmentation and prediction framework can achieve higher accuracy in medical image synthesizing for cross-modality conversion.Introduction
The combination of the multi-modality medical image as a reference or guidance for diagnosis and treatment has been developed successfully in many clinical applications. For example, PET/MRI therapy requires the combination of CT images to obtain a complete map of human tissue for attenuation correction1,2. Similarly, focused ultrasound surgery needs to be simultaneously combined with MRI as treatment guidance and CT measurements of bone structural heterogeneity as a reference for aberration correction3.Some studies have proven that deep learning neural networks, using the single modality of MRI which has the advantages of soft tissue delineation accuracy4, can generate a realistic rendering of the bone structure of CT5, thus reducing the acquisition times of multi-modality data and preliminary registration work6,7, which not only can save expenses, but also reduce CT ionizing radiation harm to patients, and improve patients’ experience 8-10.
Previous studies have attempted to achieve cross-modality translation using 2D, pseudo-3D, or 3D training strategies on various types of MRI11-16. Among them, the UTE (ultra-short-time-echo) MRI that provides high contrast and more information of bone tissue compared with other MRI sequences17,18, have been proven to be more suitable for synthesizing CT images2. However, the framework using only pixel prediction in previous work may suffer from complicated pre-processing steps.
We proposed a new processing framework combining an image segmentation task using the TABS19 model and a prediction task using Residual Attention U-net20,21 to achieve this multi-modality and multi-task image reconstruction on the 3D-patch scale. The framework that ensures the continuity of slices between 3D images and works on a very limited dataset, makes full use of the functional advantages of each model, and achieves better performance in the reconstruction.
Methods
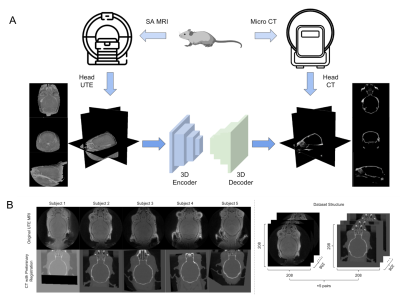
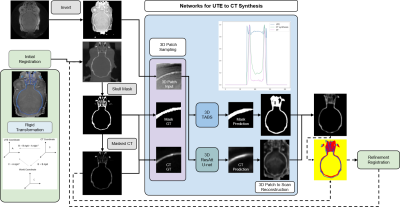
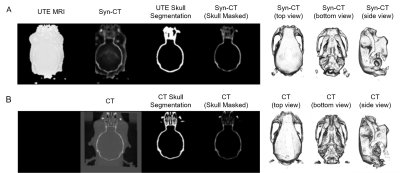
In the study, five pairs of mouse brain image were used, including UTE-MRI showing brain tissue and corresponding CT images emphasizing skull structure, as shown in Figure 1b.The data pre-processing stage was carried out according to the pipeline in Figure 2. The CT image information was projected to the UTE MRI space using rigid transformation. After the preliminary registration, the skull mask of CT was labeled by setting a threshold value to remove background information. The masked CT containing only skull structures of clinical interest was retained as the image target. Then, 32x32x32-3D-patches are extracted as the model input of double paths 3D neural networks. The synthetic CT was then reconstructed sequentially and used as a reference for the refinement registration.
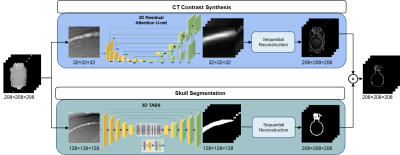
The framework of image translation is divided into two paths: the segmentation of the skull region and the prediction of pixels within the region. In Figure3, for the regression of bone pixel value, the center of patches was required to fall within the skull mask region, while for the segmentation, more background information was taken into account.
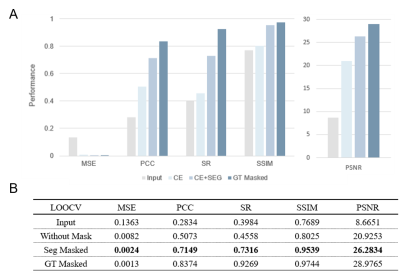
Due to the limited subjects, we adopted the training method of leaving-one-out of cross-validation (sub1 was cropped into 208×208×110 because part of the image was covered in the CT acquisition process). By averaging the evaluation results of model pairs, we proved the universality of our framework. The results are shown in Figure 4. Figure 5. shows a visualization of the synthesized CT and the corresponding ground truth. The synthesized image is highly similar to the real CT, reflecting the application potential of this processing framework for clinical cross-modality image conversion.
Results
Our 3D segmentation and pixel prediction processing framework was validated on UTE MRI to CT images of mouse brain data. All of the evaluation metrics show a significant improvement over the original UTE MRI, including Pearson Correlation Coefficient (2.5 times increased from 0.2834 to 0.7149) and Spearman (2 times increased from 0.3984 to 0.7316) average across the cross-validation.Discussion
The capability of image prediction of deep learning has been proven in cross-modality image synthesis. We find that the addition of image segmentation into the processing framework can more accurately capture the information of CT skull structure, implementing a specific area of the image reconstruction is much more effective than the global image reconstruction, and the result could be significantly improved.The challenge of this experiment is using only five subjects to achieve reliable cross-modality transformation through 3D-patch-level training without using other data augmentation. Samples extraction at the 3D-patch-level can theoretically obtain (image-size - 2 × patch-size)3 different position samples, which solves the problem of the limited dataset. At the same time, the small size 3D-patch effectively reduces the huge burden of 3D model parameters.
It was found in the process that 3D training with a larger patch size could enable the model to capture more information, thus improving the ability of model segmentation and reconstruction. But this requires stronger hardware performance and larger GPU memory.
Conclusion
Our work proposes an effective new framework for cross-modality image conversion from UTE MRI to CT and a training strategy on very limited dataset. The combination of image segmentation and prediction framework, by making model reconstruction images within the designated area, can better and faster capture the information of different modalities, to more effectively achieve the cross-modality conversion. In addition, the training strategy at the 3D-patch-level can effectively solve the difficulties of 3D global training and the deficiency of training datasets.Acknowledgements
This study was performed at the Zuckerman Mind Brain Behavior Institute at Columbia University and Columbia MR Research Center site.References
1. Wagenknecht, G., Kaiser, H.-J., Mottaghy, F.M., Herzog, H.J.M.r.m.i.p., biology & medicine. MRI for attenuation correction in PET: methods and challenges. 26, 99-113 (2013).
2. Gong, K., et al. Attenuation correction using deep Learning and integrated UTE/multi-echo Dixon sequence: evaluation in amyloid and tau PET imaging. 48, 1351-1361 (2021).
3. Vyas, U., Kaye, E. & Pauly, K.B.J.M.p. Transcranial phase aberration correction using beam simulations and MR‐ARFI. 41, 032901 (2014).
4. Prabhakar, R., et al. Comparison of computed tomography and magnetic resonance based target volume in brain tumors. 3, 121 (2007).
5. Karlsson, M., Karlsson, M.G., Nyholm, T., Amies, C. & Zackrisson, B.J.I.J.o.R.O.B.P. Dedicated magnetic resonance imaging in the radiotherapy clinic. 74, 644-651 (2009).
6. Edmund, J.M. & Nyholm, T.J.R.O. A review of substitute CT generation for MRI-only radiation therapy. 12, 1-15 (2017).
7. Beavis, A., Gibbs, P., Dealey, R. & Whitton, V.J.T.B.j.o.r. Radiotherapy treatment planning of brain tumours using MRI alone. 71, 544-548 (1998).
8. Oulbacha, R. & Kadoury, S. MRI to CT Synthesis of the Lumbar Spine from a Pseudo-3D Cycle GAN. in 2020 IEEE 17th international symposium on biomedical imaging (ISBI) 1784-1787 (IEEE, 2020).
9. Nie, D., Cao, X., Gao, Y., Wang, L. & Shen, D. Estimating CT image from MRI data using 3D fully convolutional networks. in Deep Learning and Data Labeling for Medical Applications 170-178 (Springer, 2016).
10. Hyun, C.M., et al. Deep learning for undersampled MRI reconstruction. 63, 135007 (2018).
11. Wang, Y., Liu, C., Zhang, X. & Deng, W. Synthetic CT generation based on T2 weighted MRI of nasopharyngeal carcinoma (NPC) using a deep convolutional neural network (DCNN). Front Oncol 2019; 9: 1333. (2019).
12. Han, X.J.M.p. MR‐based synthetic CT generation using a deep convolutional neural network method. 44, 1408-1419 (2017).
13. Shen, D., Wu, G. & Suk, H.-I.J.A.r.o.b.e. Deep learning in medical image analysis. 19, 221 (2017).
14. Wolterink, J.M., et al. Deep MR to CT synthesis using unpaired data. in International workshop on simulation and synthesis in medical imaging 14-23 (Springer, 2017).
15. Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. in International Conference on Medical image computing and computer-assisted intervention 234-241 (Springer, 2015).
16. Nie, D., et al. Medical image synthesis with context-aware generative adversarial networks. in International conference on medical image computing and computer-assisted intervention 417-425 (Springer, 2017).
17. Chang, E.Y., Du, J. & Chung, C.B.J.J.o.m.r.i. UTE imaging in the musculoskeletal system. 41, 870-883 (2015).
18. Robson, M.D., Gatehouse, P.D., Bydder, M. & Bydder, G.M.J.J.o.c.a.t. Magnetic resonance: an introduction to ultrashort TE (UTE) imaging. 27, 825-846 (2003).
19. Rao, V.M., et al. Improving Across-Dataset Brain Tissue Segmentation Using Transformer. (2022).
20. Wang, F., et al. Residual attention network for image classification. in Proceedings of the IEEE conference on computer vision and pattern recognition 3156-3164 (2017).
21. Wu, E., et al. CU-Net: A Completely Complex U-Net for MR k-space Signal Processing. in Proceedings of International Society of Magnetic Resonance in Medicine (2021).
Figures