5085
Repeatability of ProMyoT1: an open-source inversion recovery myocardial T1 mapping sequence for fast prototyping
Andreia S Gaspar1, Nuno A Silva2, António M Ferreira3,4, and Rita G Nunes1
1Institute for Systems and Robotics - Lisboa and Department of Bioengineering, Instituto Superior Técnico – Universidade de Lisboa, Lisbon, Portugal, 2Hospital da Luz Learning Health, Luz Saúde, Lisbon, Portugal, 3Serviço de Cardiologia, Hospital de Santa Cruz, Centro Hospitalar Lisboa Ocidental, Lisbon, Portugal, 4Unidade de Imagiologia Cardíaca Avançada, Hospital da Luz, Lisbon, Portugal
1Institute for Systems and Robotics - Lisboa and Department of Bioengineering, Instituto Superior Técnico – Universidade de Lisboa, Lisbon, Portugal, 2Hospital da Luz Learning Health, Luz Saúde, Lisbon, Portugal, 3Serviço de Cardiologia, Hospital de Santa Cruz, Centro Hospitalar Lisboa Ocidental, Lisbon, Portugal, 4Unidade de Imagiologia Cardíaca Avançada, Hospital da Luz, Lisbon, Portugal
Synopsis
Keywords: Quantitative Imaging, Heart, T1 mapping
The open-source method Prototype of Myocardial T1 mapping (ProMyoT1) created with Pulseq was tested in vivo in healthy subjects to evaluate its repeatability and compare its performance with that of the clinical MOLLI method. ProMyoT1 provided myocardium T1 values similar to those obtained with MOLLI. The precision was better for MOLLI, but the repeatability measures (Coefficient of Variation and Repeatability Coefficient) were similar in both methods. The developed ProMyoT1 method was shown to be reliable and should improve the accessibility to T1 mapping, while also enabling fast sequence prototyping so that further improvements can easily be tested.Introduction
T1 mapping has the potential to provide biomarkers of tissue disease in the heart.1 However, T1 variability due to differences in the methodology implemented by each vendor has resulted in the need to establish normative values for each imaging center. Standardization would be desirable to enable cross-site comparisons but would typically require access to vendor propriety codes, kept as black-boxes for most users. The solution is to have an open-source method that does not depend on the scanner nor the vendor. It was shown that a vendor-neutral sequence methodology may reduce sequence variations2, and Pulseq was already used to scan the same T1 and T2 mapping sequences in 1.5T and 3T scanners.3The aim of this work was to evaluate the repeatability of the recently proposed Prototype of Myocardial T1 mapping (ProMyoT1)4 method in vivo and to compare it with the standard MOLLI sequence.
Methods
ProMyoT14 is an inversion-recovery sequence using a balanced steady-state free precession (bSSFP) readout, with inversion and triggering schemes based on the MOLLI sequence, developed in Pulseq5, and available at https://github.com/ANG13/ProMyoT1. The sequence was tested with following parameters: TR/TE=3.04/1.52 ms, bSSFP readout after a linear ramp-up of 11 pulses, a sinc RF excitation pulse (490 μs and Time-Bandwidth Product of 1.5) with flip angle of 35º, slice thickness of 8 mm, 354×327 mm2 field-of-view (FOV), 256×144 matrix size, and bandwidth per pixel of 808 Hz/pixel. Physiologic triggering with a 5(3)3 scheme, with a trigger delay set for each volunteer. Each bSSFP readout was obtained with an acceleration factor of 2, with 24 lines acquired in the center of k-space, which were used as autocalibration signal in GeneRalized Autocalibrating Partial Parallel Acquisition (GRAPPA).6 Clinical MOLLI was also obtained with the same parameters except TR/TE=3.2/1.6 ms, and bandwidth of 1002 Hz/pixel.ProMyoT1 and MOLLI sequences were acquired in 21 healthy volunteers (11M, 10F), ages from 26 to 62 years (39±11 years) in a Siemens Aera 1.5 T scanner with an 18-channel body coil. For 18 volunteers both sequences were performed two times, to study its repeatability. Repeatability coefficient (RC) for a sample with n=18 subjects and m=2 independent measurement repetitions were calculated according with 7. Bland-Altman analysis was also performed to compare T1 estimates between the first and second measurements of both sequences, with Wilcoxon signed-rank test. The within-subject coefficient of variation (CV) given by the ratio of the within-subject standard deviation (SD) to its mean was also computed.
Offline reconstruction was performed for both methods. The reconstructed matrix size was 256×218. A Tukey filter with parameter 0.5 was applied. Adaptative coil combination was performed in order to obtain complex images used for Phase Sensitive Inversion Recovery (PSIR) signal that was used for T1 estimation.8 T1 estimation fitting the 3-parameter model, using the function lsqcurve, was performed offline using Matlab.
The study was approved by the local ethical board committee and written informed consent was obtained for each subject.
Results
T1 weighted (T1w) ProMyoT1 and MOLLI images (first TI) for one volunteer and respective estimated T1 maps are presented in Fig.1. The Bland-Altman analysis (Fig. 2) shows that T1 estimates were similar between ProMyoT1 and MOLLI (T1MOLLI=1004±33 ms vs T1ProMyoT1=998±52 ms), with a mean difference of -17 ms (p-value=0.20) and CI (95%)=[-86 51] ms. The segmental distribution of T1 values according with American Heart Association 16-segments model for both methods is presented in Fig. 3, where we can see that they are similar for base and middle slices, although SD for ProMyoT1 is higher. The apex slices present higher T1 values for ProMyoT1.Repeatability Bland-Altman analysis comparing scan 1 and 2 for MOLLI and ProMyoT1 is shown in Fig. 4. For MOLLI there is a mean difference of -5.9 ms (p-value=0.41) and CI (95%)=[-24 36] ms. For ProMyoT1 there is a mean difference of 5.8 ms (p-value=0.31) and CI (95%)=[-43 32] ms, with an outlier with mean T1 difference of -80 ms. Table 1 shows the values of RV, and CV for both methods, showing slightly higher values for both in ProMyoT1.
Discussion
ProMyoT1 allowed to measure myocardium T1 values similar to the clinically used MOLLI. The higher SD for ProMyoT1 can be related with higher noise present in the T1w. This can be especially problematic for apex slices, which are frequently more prone to artefacts, leading to increased T1 and mean SD ProMyoT1 estimates. Repeatability measurements were similar for both methods, showing that ProMyoT1 repeatability was not affected by its increased mean SD.Conclusion
The results presented show that the open-source sequence ProMyoT1 can be used for T1 mapping in vivo with similar results to the clinically used MOLLI method. This increases the accessibility of T1 mapping in centers that would not be able to access the vendor package that include such sequences, allowing at the same time for a base sequence from which further improvements can easily be tested. Further work includes testing the method in different scanners (e.g. 3.0T) and centers in order to study its reproducibility.Acknowledgements
Fundação para a Ciência e a Tecnologia (SFRH/BD/120006/2016; PTDC-EMD/EMD/29686/2017; UID/EEA/50009/2019; UID/EEA/50009/2020) e Programa Operacional Regional de Lisboa 2020 (LISBOA-01-0145-FEDER-029686).References
- Messroghli DR, Greiser A, Fröhlich M, Dietz R, Schulz-Menger J. Optimization and validation of a fully-integrated pulse sequence for modified look-locker inversion-recovery (MOLLI) T1 mapping of the heart. J Magn Reson Imaging. 2007;26(4):1081-1086.
- Karakuzu A, Biswas L, Cohen-Adad J, Stikov N. Vendor-neutral sequences and fully transparent workflows improve inter-vendor reproducibility of quantitative MRI. Magn Reson Med. 2022;88(3):1212–28.
- Tong G, Gaspar AS, Qian E, et al. A framework for validating open-source pulse sequences. Magn Reson Imaging. 2021;87:7-18.
- Gaspar AS, Silva NA, Nunes RG. ProMyoT1: Open-source Inversion recovery myocardial T1 mapping sequence for fast prototyping. Proc. of Annual Meeting ISMRM 2021, Virtual Meeting, 2021.
- Layton KJ, Kroboth S, Jia F, et al. Pulseq: A rapid and hardware-independent pulse sequence prototyping framework. Magn Reson Med. 2017;77:1544-1552.
- Griswold MA, Jakob PM, Heidemann RM, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med.2002;47:1202-1210.
- Ye S, Lim JY, Huang W. Statistical considerations for repeatability and reproducibility of quantitative imaging biomarkers. BJR|Open. 2022;4(1):20210083.
- Xue H, Greiser A, Zuehlsdorff S, et al. Phase-sensitive inversion recovery for myocardial T1 mapping with motion correction and parametric fitting. Magn Reson Med. 2013;69.
Figures
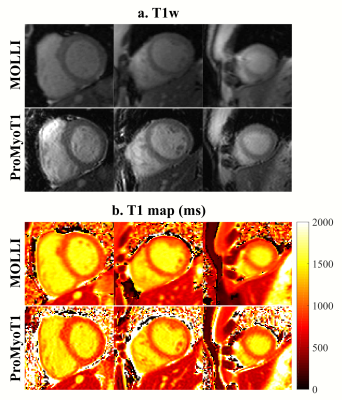
Figure 1: T1 weighted (T1w) (a.) and respective T1 map (in ms) (b.) for
base, middle and apex slices of MOLLI and ProMyoT1 in one healthy
volunteer.

Figure 2: T1 mean values over 21 volunteers and mean standard deviation (SD) (in ms) according with AHA 16 segment model for MOLLI and ProMyoT1.
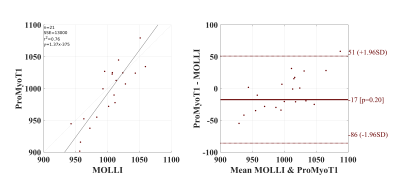
Figure 3: Bland-Altman analysis for T1 values obtained with ProMyoT1 compared with MOLLI. p-value presented are calculated with paired, two-sided Wilcoxon signed rank test for zero median of the hypothesis that the difference between the matched samples T1 values comes from a distribution whose median is zero.
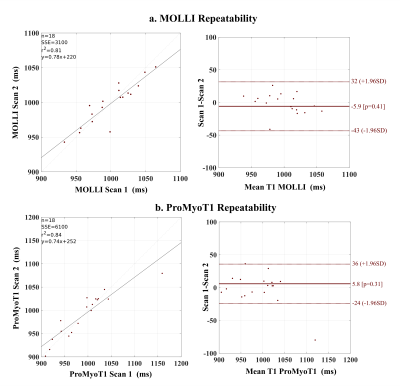
Figure 4: Repeatability of a. MOLLI, and b.ProMyoT1, on 18 volunteers. Bland-Altman analysis for T1 values comparing between scan repetitions.
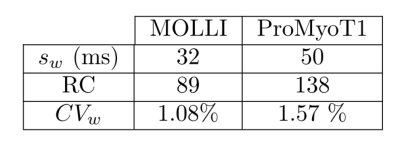
Table 1: Repeatability measures for test-retest: standard deviation within-subject (sw), Repeatability coefficient (RC), coefficient variation within-subject (CVw).
DOI: https://doi.org/10.58530/2023/5085