5041
3D Lesion Generation Model considering Anatomic Localization to Improve Object Detection in Limited Lacune Data1Department of Electrical and Electronic Engineering, Yonsei Univ., Seoul, Korea, Republic of, 2Department of Artificial Intelligence, Sejong Univ., Seoul, Korea, Republic of, 3Department of Biomedical Engineering, Yonsei Univ., Wonju, Korea, Republic of, 4Department of Neurology, Gachon Univ., Incheon, Korea, Republic of
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Data Processing, Augmentation
Recently, research on the detection of cerebral small vessel disease (CSVD) has been mainly implemented in two-stages (1st: candidate detection, 2nd: false-positive reduction). Previous studies presented the difficulty of collecting labeled data as a limitation. Here, we synthesized the lesion through 3D-DCGAN and insert it at different locations on the MR image considering anatomical localization and alpha blending to augment labeled data. Through this, the detecting architecture was simplified to a single-stage, and the precision and recall values were improved by an average of 0.2.INTRODUCTION
Cerebral small vessel disease (CSVD) is a variety of abnormalities in small vessels of the brain, including lacunes, microbleeds, and hemorrhages 1-3. CSVD remains the leading cause of death and dysfunction worldwide 4. The lacune, a type of CSVD, is defined as a small lesion resulting from the blockage of a penetrating branch of the main cerebral artery 5. Recently, many studies have been developed on the automatic detection of lesions to identify lacunes accurately 6,7. Previous studies on CSVD detection were mainly implemented in two-stages (1st: candidate detection, 2nd: false-positive reduction) 7,8. Manual labeling by radiologists is laborious, time-consuming, and subjective, pointing out the limitations of data collection 9.In this study, we synthesized the lacune patch through 3D-DCGAN 10. The synthesized lesion patches were inserted back into the MR images at various anatomical localizations where the lacune could occur. At this time, the synthesized patches were overlaid harmoniously through the alpha blending method 11. This means not only synthesizing the lesion but also augmenting labeled data in MR images. We compared the detection performance of the augmented dataset and the non-augmented dataset, and the precision and recall values were improved by an average of 0.2. Also, the proposed method has the advantage that the detection architecture could be configured as a single-stage.
METHODS
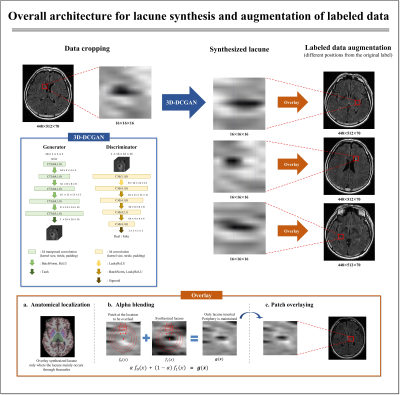
[Data augmentation]Figure 1 shows the overall architecture for augmenting labeled data. In the generator, a 16×16×16 patch including the feature map of lacune was generated through transposed convolution. After that, extract the feature map of the input data and distinguish between fake and real from the discriminator. By repeating these processes, the fake lacune approximates the probability density function (PDF) of the real lacune. To efficiently solve the binary choice problem, we applied binary cross entropy loss 12 (BCE loss) for the loss function:
$$BCE\ loss=-\frac{1}{N}\sum_{i=0}^{N}y_i\cdot\log{(\hat{y}_i)}+(1-y_i)\cdot\log{(1-\hat{y}_i)}$$
And we implemented alpha blending 11 to harmoniously overlay the synthesized lacune patch.
$$g(x)=αf_0(x)+(1-α)f_1(x)$$
where f0(x) denotes the 3D patch data at the location to be overlaid in MR images and f1(x) denotes the 3D patch data of the synthesized lacune. When overlaying the synthesized lacune, the periphery should hold f0(x) and the center should have f1(x). Therefore, the synthesized lacune images were harmoniously overlaid by setting the alpha weight value according to the distance from the center point of the data (Fig. 1).
[Anatomical localization]
About 80% of lacunes occur in the basal ganglia, especially the putamen, thalamus, and white matter of the internal capsule, pons 13. 3D segmentation of the brain MR image of each patient was performed using Freesurfer 14, and the generated lesion was randomly inserted at the anatomical localization of the lacune. Through this, labeled data was augmented considering not only the shape of the lacune but also the location.
[Detection model]
We applied Yolov5 15 as a detection model. Yolov5 consists of three parts: a backbone that extracts features, a neck that improves performance by fusion of the extracted features, and a head that converts features into bounding box parameters. With this learning pipeline, the learning speed is faster than the existing R-CNN detection models, and learns the context of the entire image.
[Dataset]
The dataset was collected at Gachon University College of Medicine Gil Hospital. FLAIR data were obtained and 76 lacunes in 52 patients were used as datasets. 50 lacunes were used for training and validation, and this is called a non-augmented dataset. In contrast, an augmented dataset was constructed by overlaying 50 synthesized lacunes to the patient MR image used in the train. The remaining 26 lacunes were used as the test set, and the detection performance was compared on the non-augmented and augmented datasets.
RESULTS
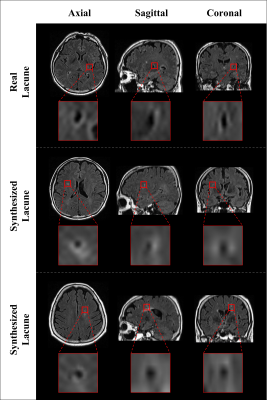
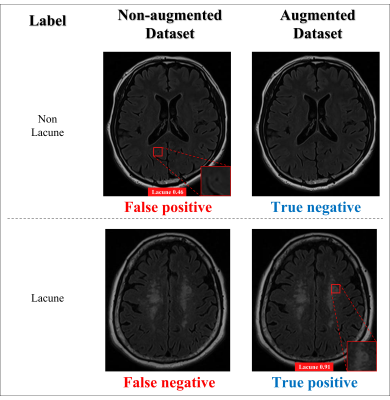
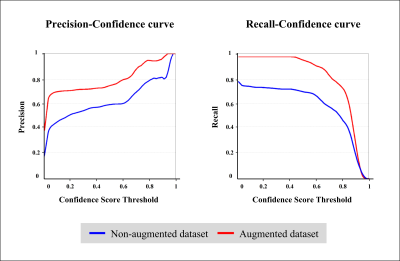
The results of the real lacune and the synthesized lacune were shown as axial, sagittal, and coronal in Fig. 2. Figure 3 compares the detection results of the non-augmented dataset and the augmented dataset on the same data. In the lacune data, the non-augmented dataset failed to detect lacunes with a confidence score of less than 0.25, resulting in false negatives. On the contrary, in the augmented dataset, it was detected with a high confidence score of 0.91. In Figure 4, between the confidence scores of 0.2 and 0.8, there is an average improvement of about 0.2 in both precision and recall.DISCUSSION AND CONCLUSION
In this work, we implemented 3D-DCGAN to improve the detection performance of lesions. In addition, anatomical localization of the lesion was considered using Freesurfer, and the synthesized lesion was harmoniously overlaid through alpha blending. Through this, lesions were synthesized at different locations in MR images and completely new labeled data were obtained. This gave an advantage to the detection model that adjusts the position or size of the anchor box through training, and the detection performance was also improved. However, the size and shape of the lesion were synthesized in various ways, and the alpha blending has a limitation in that the alpha weight value for each area is fixed. The future research direction is to study a generative model that synthesizes lesions while preserving peripheral global information without such an image blending technique.Acknowledgements
This research was supported by the Brain Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (2018M3C7A1056884) and (NRF-2019R1A2C1090635).References
1. Cuadrado-Godia E, Dwivedi P, Sharma S, et al. Cerebral small vessel disease: a review focusing on pathophysiology, biomarkers, and machine learning strategies. Journal of stroke. 2018;20(3):302.
2. Pantoni L. Cerebral small vessel disease: from pathogenesis and clinical characteristics to therapeutic challenges. The Lancet Neurology. 2010;9(7):689-701.
3. Alistair D G. Hypertensive cerebral small vessel disease and stroke. Brain pathology. 2002;12(3):358-370.
4. Lee W J, Chou K H, Lee P L, et al. Cerebral small vessel disease phenotype and 5-year mortality in asymptomatic middle-to-old aged individuals. Scientific Reports. 2021;11(1):1-10.
5. Ling Y, Chabriat H. Incident cerebral lacunes: a review. Journal of Cerebral Blood Flow & Metabolism. 2020;40(5):909-921.
6. Ghafoorian M, Karssemeijer N, Heskes T, et al. Deep multi-scale location-aware 3D convolutional neural networks for automated detection of lacunes of presumed vascular origin. NeuroImage: Clinical. 2017;14:391-399.
7. Al-Masni M A, Kim W R, Kim E Y, et al. 3D multi-scale residual network toward lacunar infarcts identification from MR images with minimal user intervention. IEEE Access. 2021;9:11787-11797.
8. Myung M J, Lee K M, Kim H G,et al. Novel Approaches to detection of cerebral microbleeds: single deep learning model to achieve a balanced performance. Journal of Stroke and Cerebrovascular Diseases. 2021;30(9):105886.
9. Al-Masni M A, Kim W R, Kim E Y, et al. Automated detection of cerebral microbleeds in MR images: A two-stage deep learning approach. NeuroImage: Clinical. 2020;28:102464.
10. Momeni S, Fazlollahi A, Lebrat L, et al. Generative Model of Brain Microbleeds for MRI Detection of Vascular Marker of Neurodegenerative Diseases. Frontiers in Neuroscience. 2021;15.
11. Guttman M A, Lederman R J, Sorger J M, et al. Real-time volume rendered MRI for interventional guidance. Journal of Cardiovascular Magnetic Resonance. 2002;4(4):431-442.
12. Ruby U, Theerthagiri P, Jacob J, et al. Binary cross entropy with deep learning technique for image classification. Int J Adv Trends Comput Sci Eng. 2020;9(10).
13. Elsevier Health Sciences. Stroke e-book: Pathophysiology, diagnosis, and management. Philadelphia: Elsevier Health Sciences; 2021.
14. Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774-781.
15. Dipu N M, Shohan S A, and Salam K M A. Deep learning based brain tumor detection and classification. In 2021 International Conference on Intelligent Technologies (CONIT). IEEE. 2021:1-6.
Figures