5040
Amyloid-Beta Axial Plane PET Synthesis from Structural MRI: An Image Translation Approach for Screening Alzheimer’s Disease1Biomedical Engineering, Schulich School of Engineering, University of Calgary, Calgary, AB, Canada, 2Electrical & Software Engineering, Schulich School of Engineering, University of Calgary, Calgary, AB, Canada, 3Hotchkiss Brain Institute, Cumming School of Medicine, University of Calgary, Calgary, AB, Canada, 4Department of Radiology, Cumming School of Medicine, University of Calgary, Calgary, AB, Canada
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Alzheimer's Disease, MRI, PET, Image Translation
In this work, an image translation model is implemented to produce synthetic amyloid-beta PET images from structural MRI that are quantitatively accurate. Image pairs of amyloid-beta PET and structural MRI were used to train the model. We found that the synthetic PET images could be produced with a high degree of similarity to truth in terms of shape, contrast and overall high SSIM and PSNR. This work demonstrates that performing structural to quantitative image translation is feasible to enable the access amyloid-beta information from only MRI.Introduction
Alzheimer’s Disease (AD) is the most prevalent cause of dementia, with an estimated economic burden of $305 Billion in 2020 in the US.1 By the time that AD is clinically diagnosed, the patient is already experiencing neuronal loss and brain atrophy. Amyloid-beta is a key molecule that is widely believed to be a root cause of AD pathophysiology that starts aggregating within the brain before clinical symptoms manifest.2,3Over the last 15-years, it has become possible to image amyloid-beta with the use of positron emission tomography (PET) tracers4 however, PET imaging has some disadvantages such as: cost ($5000-to-$8000 per scan), invasiveness as it requires the injection of a radiotracer leading to radiation exposure, and the tracers are not available in many jurisdictions5 these disadvantages limit the use for screening and early onset detection.
Structural MRI is about 10-times cheaper than PET ($500-to-$800) and can aid to assess AD by examining structural changes and atrophy.6 MRI has limited ability to provide molecular information, but image translation algorithms might be used to compute synthetic amyloid-beta PET images from structural MRI. The translation is possible due to the relationships between amyloid-beta burden and brain atrophy.7,8
Image translation is an advanced form of machine learning that aims to render one image type from another, enabling the access to images that might not otherwise be captured.9 These algorithms are commonly implemented using Convolutional Neural Networks (CNN)10 with Conditional Generative Adversarial Networks (cGAN)11 as they excel in generating realistic images.12 cGANs require pairs of images that share mutual information.13
This work demonstrates a pipeline using cGAN to generate quantitatively accurate synthetic amyloid-beta PET images from structural MRI images.
Methods
The Open Access of Imaging Studies (OASIS-3)14 dataset provided 929 subjects with pairs of T1-weighted MRI and amyloid-beta PET images, from these 609 are cognitively normal (CN) subjects and 489 at different stages of cognitive decline. The MRI images were obtained with three different scanners: Siemens Biograph mMR 3T, Siemens Trio Tim 3T, and Siemens Sonata 1.5T with a resolution of 1mm×1mm×1mm for the 3T scanners and 1.2mm×1.2mm×1.2mm for the 1.5T scanners. The amyloid-beta PET images were obtained with two scanners: Siemens Biograph 40 PET/CT and Siemens ECAT 962 using Pittsburgh Compound B15 as a radiotracer with an administered dose that ranged between 6 – 20 millicuries (mCI) and a 60-minute dynamic PET scan where Standard Uptake Value Ratio (SUVr) was obtained using the PET Unified Pipeline.16For preprocessing, the dynamic PET images were converted to static by summing the last 30 minutes of the tracer passage, then PET images where brain extracted to eliminate scattering outside the head generated by the radiotracer, the MRI images where preprocessed with Freesurfer that produced the brain extractions and both PET and MRI images were co-registered to the Montreal Neurological Institute (MNI) 1mm template using a composite transform that comprised translation and affine registration using Advanced Normalization Tools (ANTs), and split them in the axial plane.
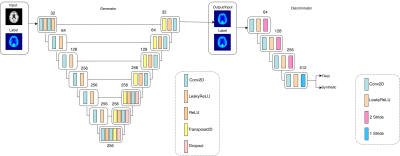
The cGAN architecture shown in Figure 1 was implemented to conduct image translation using the pytorch deep learning library.17 A custom loss function was implemented to mask the image during training and only penalize information inside of the brain. The model was trained with 550 subjects and stratified with equal proportion of females and males and different levels of cognitive impairment levels.
The model was tested with 335 unseen MRI images to generate synthetic PET images that were evaluated with their paired true PET images by examining the quality of individual cases and population with Structural Similarity Index (SSIM)18 and Peak Signal-to-Noise Ratio (PSNR).19
Results
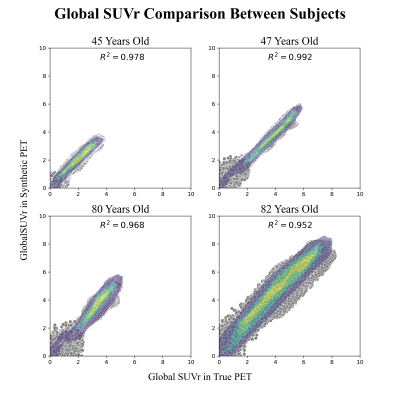
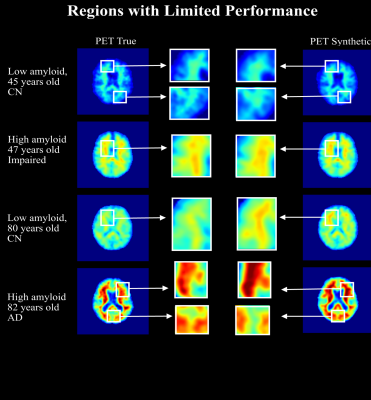
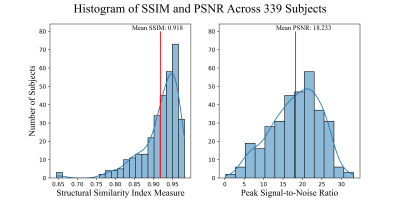
Four subjects were selected as examples showing how the model is able to produce high quality synthetic amyloid-beta PET images for both low and high amyloid cases, in both younger and older participants. Figure 2, contains inputs, truth cases, synthesized images and difference maps. The SUVR comparison in Figure 3, reports R2>0.95 for the four cases implying that the model is able to produce quantitative accurate synthetic amyloid-beta PET images. Figure 4 shows magnifications focusing on regions with limited performance.Figure 5 shows a high overall SSIM and PSNR meaning that the model generalizes well to the population of unseen images.
Discussion & Conclusion
The future work for this project will include improving the performance, particularly of the small discrepancies seen in Figure 4, by exploring different ways to of assessing the quantification during training. This is a known problem in generative models as most of the implementations and metrics focus on generating relative images by normalizing the pixel values disregarding the quantification of the contrast.20,21 Future work will also include developing a 3D model while targeting the quantification problem.Here-in, a pipeline to generate high-quality synthetic amyloid-beta PET images from structural MRI was implemented, the results are encouraging with inspective images having a high degree of similarity and population wide metrics shows SSIM>0.9. The method provides a non-invasive tool for early AD screening.
Acknowledgements
The authors would like to thank the University of Calgary, in particular the Schulich School of Engineering and Departments of Biomedical Engineering and Electrical & Software Engineering; the Cumming School of Medicine and the Departments of Radiology and Clinical Neurosciences; as well as the Hotchkiss Brain Institute, Research Computing Services and the Digital Alliance of Canada for providing resources. The authors would like to thank the Open Access of Imaging Studies Team for making the data available. FV – is funded in part through the Alberta Graduate Excellence Scholarship. JA – is funded in part from a graduate scholarship from the Natural Sciences and Engineering Research Council Brain Create. AE – was funded in part from the Biomedical Engineering Summer Studentship Program. MEM acknowledges support from Start-up funding at UCalgary and a Natural Sciences and Engineering Research Council Discovery Grant (RGPIN-03552) and Early Career Researcher Supplement (DGECR-00124). This work was made possible through a generous donation by Jim Gwynne.
References
1. Wong, W., Economic burden of Alzheimer disease and managed care considerations. Am J Manag Care, 2020. 26(8 Suppl): p. S177-S183.
2. Hampel, H., et al., The Amyloid-β Pathway in Alzheimer’s Disease. Molecular Psychiatry, 2021. 26(10): p. 5481-5503.
3. Chen, G.-f., et al., Amyloid beta: structure, biology and structure-based therapeutic development. Acta Pharmacologica Sinica, 2017. 38(9): p. 1205-1235.
4. Kolanko, M.A., et al., Amyloid PET imaging in clinical practice. Pract Neurol, 2020. 20(6): p. 451-462.
5. Marti-Climent, J.M., et al., Effective dose estimation for oncological and neurological PET/CT procedures. EJNMMI Res, 2017. 7(1): p. 37.
6. Khan, T.K., Biomarkers in Alzheimer's disease. 2016. 1 online resource (278 pages).
7. Tosun, D., et al., Spatial patterns of brain amyloid-beta burden and atrophy rate associations in mild cognitive impairment. Brain, 2011. 134(Pt 4): p. 1077-88.
8. Chetelat, G., et al., Relationship between atrophy and beta-amyloid deposition in Alzheimer disease. Ann Neurol, 2010. 67(3): p. 317-24.
9. Pang, Y., et al. Image-to-Image Translation: Methods and Applications. 2021. arXiv:2101.08629.
10. Yamashita, R., et al., Convolutional neural networks: an overview and application in radiology. Insights into Imaging, 2018. 9(4): p. 611-629.
11. Isola, P., et al., Image-to-Image Translation with Conditional Adversarial Networks. 30th Ieee Conference on Computer Vision and Pattern Recognition (Cvpr 2017), 2017: p. 5967-5976.
12. Goodfellow, I.J., et al. Generative Adversarial Networks. 2014. arXiv:1406.2661.
13. Yi, X., E. Walia, and P. Babyn Generative Adversarial Network in Medical Imaging: A Review. 2018. arXiv:1809.07294.
14. LaMontagne, P.J., et al., OASIS-3: Longitudinal Neuroimaging, Clinical, and Cognitive Dataset for Normal Aging and Alzheimer Disease. medRxiv, 2019.
15. Yamin, G. and D.B. Teplow, Pittsburgh Compound-B (PiB) binds amyloid beta-protein protofibrils. J Neurochem, 2017. 140(2): p. 210-215.
16. Washington, U.o. PET Unified Pipeline. 2019; Available from: https://github.com/ysu001/PUP.
17. Paszke, A., et al., PyTorch: An Imperative Style, High-Performance Deep Learning Library, in Advances in Neural Information Processing Systems 32, H. Wallach, et al., Editors. 2019, Curran Associates Inc. p. 8024--8035.
18. Nilsson, J. and T. Akenine-Möller Understanding SSIM. 2020. arXiv:2006.13846.
19. Fardo, F.A., et al. A Formal Evaluation of PSNR as Quality Measurement Parameter for Image Segmentation Algorithms. 2016. arXiv:1605.07116.
20. Sikka, A., et al. MRI to PET Cross-Modality Translation using Globally and Locally Aware GAN (GLA-GAN) for Multi-Modal Diagnosis of Alzheimer's Disease. 2021. arXiv:2108.02160.
21. Zhang, J., et al., BPGAN: Brain PET synthesis from MRI using generative adversarial network for multi-modal Alzheimer's disease diagnosis. Comput Methods Programs Biomed, 2022. 217: p. 106676.
Figures