5037
Synthesising MRIs from CTs to Improve Stroke Treatment Using Deep Learning1Auckland Bioengineering Institute, University of Auckland, Auckland, New Zealand, 2Faculty of Medical and Health Sciences, University of Auckland, Auckland, New Zealand, 3Mātai Medical Research Institute, Tairāwhiti-Gisborne, New Zealand
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Stroke, Image Synthesis
MRI holds an important role in diagnosing brain conditions, however, many patients do not receive an MRI before their diagnosis and onset of treatment. We propose to use deep learning to generate an MRI from a patient's CT and have implemented multiple models to compare their results. Using CT/MRI pairs from 181 stroke patients, we use mutiple deep learning models to generate MRI from the CT images. The model produces high quality images and accurately translates lesions onto the target image.
Introduction
MRI is important in diagnosing stroke as it has superior soft tissue contrast compared to other modalities and provides valuable information for accurate diagnosis and for better management and prediction of outcomes1. CT is the modality of choice for screening brain conditions as it has the benefit of being quicker, more comfortable, and cheaper than other modalities of imaging2. However, on top of having the better soft tissue contrast of MRIs, most of the commonly used brain atlases are constructed from MRIs which means that an MRI of the patient is preferable for comparison3. A solution is to synthesise the unobtained MRI from the obtained CT.Image generation using deep learning is a growing field with many applications. Unet4 is a convolutional neural network developed for the segmentation of biomedical images and more recently has been used to show the plausibility of medical image synthesis5-7. However, this usually focuses on generating CT scans from MRIs. The benefit of UNet is that it works with paired data, meaning it can learn the mapping from CT to MRI for each patient. On the other hand, generative adversarial networks8 have gained extreme popularity in image synthesis however work by making the output fit into the target domain distribution, leading to high-quality images that may have hallucinated features9.
Method
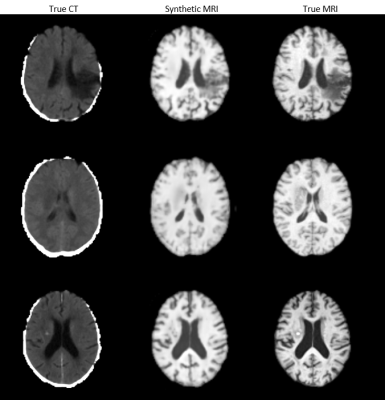
The dataset's quality is of utmost importance in training a deep learning model10. Creating a robust preprocessing pipeline for the data was an essential first step. We had access to a dataset from the Auckland District Health Board, including CT and T1-weighted MRI scans of 199 patients post-stroke. The pipeline consisted of registering the CT to the MRI using linear registration11-13, registration of the MRI and CT to an atlas using linear registration, applying HD_BET14 to the MRI to extract the brain and using the extracted mask to extract the CT brain, normalising the MRI, and finally, a downsampling and cropping step. After this preprocessing step, CT/MR pairs from 181 patients remained, and a train/validation/test split of 132/24/25 was used.A five-layer UNet architecture was implemented to suit our dataset, and hyperparameters were tuned to get the optimal performance of such a model. A second model was developed based on the first where convolution blocks were added into the skip connections to implement a Unet++15 architecture. Finally, a third model was developed based on the first but with attention gates in the skip connections to implement AttentionUNet16. All implementations took in 3D data.
The original UNet model was trialed with 5 different loss functions: MAE, MSE, PSNR, SSIM, and MAE + MSE. MAE gave the best performance, so this was the loss function used for all three models - the other metrics (MSE, PSNR, and SSIM) were tracked throughout training. All models were run through 400 epochs with a learning rate of 10-3.
Results
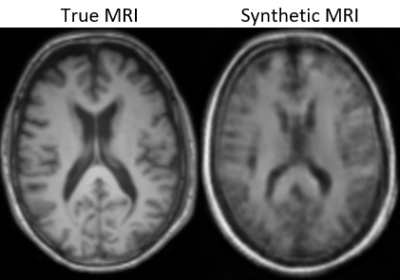
UNet with an MAE loss demonstrated the best image fidelity and quality across the test data compared to other loss functions (Figure 1). The performance of UNet, UNet++, and AttentionUNet were comparable with AttentionUNet showing a slight improvement on some of the fine details (Figure 2). The UNet model also shows potential in transferring lesions across to the synthetic MRI (Figure 3). Figure 4 shows a preliminary result from 3D-CycleGAN17,18 run on an iteration of the dataset that did not undergo brain extraction.Discussion
All three UNet models demonstrate potential for generating high-quality MRIs. Recently, transformers have gained much popularity in many deep learning19 and they have been implemented into UNet segmentation models in multiple ways20,21. Incorporating transformers into the UNet model for MRI synthesis is an avenue for future research we will explore. Previous research into medical image synthesis focused mainly on generative adversarial networks, however, these can lead to hallucinated image features such as lesions, when they are not present or the lack of lesions when they are present9. Our results show similar quality to other studies6,7, however, we propose the addition of transformers into a UNet model for synthesis to improve the results, which we are currently working on. Building up multi-center pairwise CT and MRI datasets can further improve the diversity and robustness of the image generation models.Conclusion
Synthesising MRIs from CTs could have a huge impact on the treatment of stroke. We have developed multiple UNet models that produce brain images comparable to state-of-the-art results in this area, with some slight improvement on some fine structural details. Applying this method to larger datasets holds promise in improving the diversity and robustness of the image generation model.Acknowledgements
This work was partially supported by the Health Research Council of New Zealand [grant number 21/144].References
1. Kim, B.J., et al., Magnetic Resonance Imaging in Acute Ischemic Stroke Treatment. Journal of Stroke, 2014. 16(3): p. 131.
2. Buttram, S.D., et al., Computed tomography vs magnetic resonance imaging for identifying acute lesions in pediatric traumatic brain injury. Hosp Pediatr, 2015. 5(2): p. 79-84.
3. Nowinski, W.L., Usefulness of brain atlases in neuroradiology: Current status and future potential. Neuroradiol J, 2016. 29(4): p. 260-8.
4. Ronneberger, O., P. Fischer, and T. Brox, U-Net: Convolutional Networks for Biomedical Image Segmentation, in Lecture Notes in Computer Science. 2015, Springer International Publishing. p. 234-241.
5. Li, Y., et al., Comparison of Supervised and Unsupervised Deep Learning Methods for Medical Image Synthesis between Computed Tomography and Magnetic Resonance Images. BioMed Research International, 2020. 2020: p. 1-9.
6. Li, W., et al., Magnetic resonance image (MRI) synthesis from brain computed tomography (CT) images based on deep learning methods for magnetic resonance (MR)-guided radiotherapy. Quantitative Imaging in Medicine and Surgery, 2020. 10(6): p. 1223-1236.
7. Kalantar, R., et al., CT-Based Pelvic T1-Weighted MR Image Synthesis Using UNet, UNet++ and Cycle-Consistent Generative Adversarial Network (Cycle-GAN). Front Oncol, 2021. 11: p. 665807.
8. Goodfellow, I., et al., Generative Adversarial Networks. Advances in Neural Information Processing Systems, 2014. 3.
9. Cohen, J.P., M. Luck, and S. Honari, Distribution Matching Losses Can Hallucinate Features in Medical Image Translation. 2018, Springer International Publishing. p. 529-536.
10. Sessions, V. and M. Valtorta, The Effects of Data Quality on Machine Learning Algorithms. 2006. 485-498.
11. Greve, D.N. and B. Fischl, Accurate and robust brain image alignment using boundary-based registration. NeuroImage, 2009. 48(1): p. 63-72.
12. Jenkinson, M. and S. Smith, A global optimisation method for robust affine registration of brain images. Med Image Anal, 2001. 5(2): p. 143-56.
13. Jenkinson, M., et al., Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage, 2002. 17(2): p. 825-41.
14. Isensee, F., et al., Automated brain extraction of multisequence MRI using artificial neural networks. Human Brain Mapping, 2019. 40(17): p. 4952-4964.
15. Zhou, Z., et al., UNet++: A Nested U-Net Architecture for Medical Image Segmentation. 2018, Springer International Publishing. p. 3-11.
16. Oktay, O., et al., Attention U-Net: Learning Where to Look for the Pancreas. 2018.
17. Iommi, D., 3D-CycleGan-Pytorch-MedImaging. 2021.
18. Zhu, J.-Y., et al., Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. 2017. 2242-2251.
19. Vaswani, A., et al., Attention Is All You Need. 2017.
20. Petit, O., et al., U-Net Transformer: Self and Cross Attention for Medical Image Segmentation. 2021, Springer International Publishing. p. 267-276.
21. Chen, J., et al., TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. 2021.
Figures
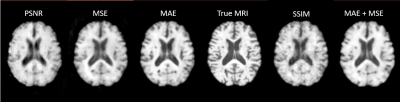
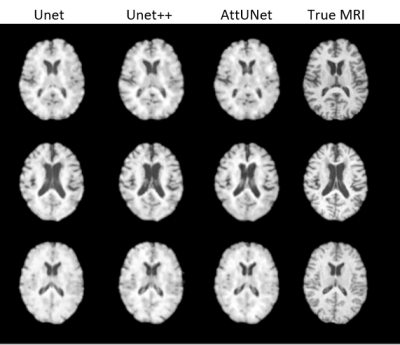
Figure 2: The Synthetic MRIs generated by UNet, UNet++, and AttUNet, alongside the True MRIs.