5036
Synthetic CT generation from different MR contrast inputs and evaluation of its quantitative accuracy1GE Healthcare, Munich, Germany, 2Department of Quantitative Biomedicine, University of Zurich, Zurich, Switzerland, 3Newcastle University and Northern Centre for Cancer Care, Newcastle upon Tyne, United Kingdom, 4Newcastle University, Newcastle upon Tyne, United Kingdom
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Radiotherapy, Synthetic CT, radiation therapy planning, Multi-task network, deep learning
MRI to synthetic CT image conversion is a problem of interest for clinical applications such as MR-radiation therapy planning, PET/MR attenuation correction, MR bone imaging. Many methods proposed for this purpose use different MR inputs. In this work, we compare the sCT generated from different 3D MR inputs, including Zero TE (ZTE), fast spin echo (CUBE), and fast spoiled gradient echo with Dixon-type fat-water separation (LAVA-Flex), using a multi-task deep learning (DL) model. We analyze the qualitative and quantitative accuracy of the generated sCT image from each input and highlight the aspects relevant for different clinical applications.Introduction
Generation of synthetic CT (sCT [HU]) from MR images is of interest for applications such as MR-only radiation therapy (RT) planning, PET/MR attenuation-correction (AC), and MR bone imaging. Different proposed methods for this problem use various image contrasts [1,2,3,4]. Each MR contrast brings a few advantages at a certain cost. In this work, we compare the sCT generated from different 3D MR inputs, including Zero TE (ZTE), fast spin echo (CUBE), and fast spoiled gradient echo with Dixon-type fat-water separation (LAVA-Flex), using a multi-task deep learning (DL) model. We analyze the qualitative and quantitative accuracy of the generated sCT image from each input and highlight the aspects relevant for different clinical applications.Methods and Materials
Patient data: MR scans were performed using a 3T, time-of-flight (TOF) Signa PET/MR scanner (GE Healthcare, Chicago, IL, USA). 52 pelvis radiation oncology patients were scanned with three different MR sequences including 3D PDw ZTE, 3D T2w CUBE, and PDw LAVA-Flex (Table-1). For all patients, accompanying CT scans were available from earlier examinations. All patient studies were approved by respective Institutional Review Boards, including signed informed consent. CT to MR registration: Each MR contrast was registered separately to the patient’s CT image using a combination of rigid and diffeomorphic dense registration algorithms developed in ITK [5]. All MR images were co-registered to the CT image for comparison of each sCT in a normalized geometric space.Deep learning based sCT computation: A 2D supervised multi-task CNN in a UNet like architecture was developed to compute sCT from ZTE and T2 MRI [6]. The same DL model was adapted to process multiple inputs for the LAVA-Flex based sCT generation. Of the available data, 47 cases were divided 80:20 for training and validation cohort. The remaining 5 cases were set apart for performance testing. The data was augmented with random flips, rotations, and arbitrary multiplicative bias to simulate MR inhomogeneity. Training was performed on 12260 slices from a total of 83 image volumes and each epoch was validated on 3583 slices from 37 image volumes. Predicted slices were reconstituted to form the whole sCT volume. sCT evaluation: We computed MAE in different tissue regions between sCT and real CT as a measure of HU value prediction accuracy. Dice similarity coefficient between CT bone region and DL predicted bone region as a measure of bone classification accuracy. The similarity of tissue and bone value probability distribution provides a qualitative impression of the overall HU value accuracy in different regions in a qualitative manner.
Result & Discussion
Visual comparison of the generated sCT images with the corresponding real CT shows the comparable bone depiction in the sCT output maintained with each different MR input contrast (Fig.1). The soft-tissue details are depicted best with Dixon input followed by similar details depicted with T2w and ZTE inputs. Fig.2 shows the comparison of HU value distribution in soft-tissue and bone regions over the entire image between real CT and sCT from different inputs. The quantitative metrics in Table-2 shows the MAE in different tissue regions in the test cases. ZTE offers the best bone accuracy followed by Dixon and T2w inputs. ZTE and Dixon inputs offer similar soft-tissue MAE on an average, but sCT with Dixon input demonstrates more accurate structural soft-tissue details depiction. ZTE image input offers fast scan advantage, Dixon input offers better soft-tissue depiction with the tradeoff of a longer scan. T2w input produces sCT of comparable appearance to the other two inputs, with an even longer scan time, but at a reduced bone value accuracy.Conclusion
We have presented a comparison of sCT generation from three different MR acquisitions. sCT generated from each MR input has similar characteristics and accuracy compared to real CT. Each input offers an advantage and allows the flexibility to generate accurate sCT images using MR input of choice dependent on the underlying application. Relevant considerations in the context of MR-only RT planning include 1) synergistic use of T2 for organs-at-risk (OAR) or tumor delineation, and 2) importance of soft-tissue contrast for positioning during RT treatment (i.e, bone and/or soft-tissue matching).Acknowledgements
This research is part of the Deep MR-only Radiation Therapy activity that has received funding from EITHealth. EIT Health is supported by the European Institute of Innovation and Technology (EIT), a body of the European Union receives support from the European Union’s Horizon 2020 Research and innovation programReferences
[1]. H.A Massa, et al, Phys. Med. Biol. 2020, Vol.65 23NT03
[2]. MF Spadea, et al, Medical Physics 2021; Vol.48 Issue.11, Pages 6537-6566
[3]. Y Li, et al, BioMed Research International, vol. 2020, Article ID 5193707, 9 pages, 2020
[4]. E Persson, et al, International Journal of Radiation Oncology, Biology, Physics, Volume 99, Issue 3, 692 - 700
[5]. B.B Avants, et al, Penn Image Computing and Science Laboratory, 2009
[6]. S Kaushik, et al, arXiv:2203.16288v2
Figures
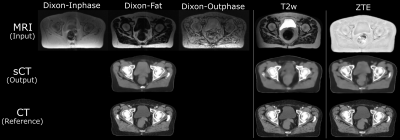
Fig.1: Pelvis sCT images generated from the proposed method maintains structural match to input MR image and bears realistic appearance of true CT. Dixon input enables an improved depiction of both soft-tissue and bone regions. sCT from ZTE and T2w inputs depict the bones with better accuracy compared to soft-tissue regions, but preserve the average difference from CT within acceptable range.
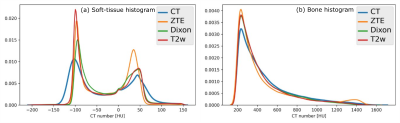
Fig.2: The probability distribution of the HU values in (a) soft-tissue region and (b) bone region for a typical pelvis image volume. The closeness of sCT distribution to real CT shows the accuracy of HU value predicted in sCT images.

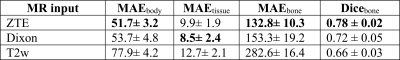
Table-2: Quantitative evaluation of the sCT output for different MR input contrast. ZTE input results in a better bone value estimation and bone classification. Dixon input provides a better soft-tissue value prediction.