5035
Prediction of NF2 Loss in Meningiomas Using T1-Weighted Contrast Enhanced MRI Generated by Deep Convolutional Generative Adversarial Networks1Electrical and Electronics Engineering, Bogazici University, Istanbul, Turkey, 2Institute of Biomedical Engineering, Bogazici University, Istanbul, Turkey, 3Health Institutes of Turkey, Istanbul, Turkey, 4Department of Medical Pathology, Acibadem University, Istanbul, Turkey, 5Brain Tumor Research Group, Acibadem University, Istanbul, Turkey, 6Department of Medical Engineering, Acibadem University, Istanbul, Turkey, 7Department of Neurosurgery, Acibadem University, Istanbul, Turkey, 8Department of Radiology, Acibadem University, Istanbul, Turkey
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence
Neurofibromatosis type 2 (NF2) gene mutations have been linked to tumorigenesis in meningiomas. This study aims to improve the prediction of NF2 loss in meningiomas using T1-weighted contrast-enhanced MRI augmented by a deep convolutional generative adversarial network (DCGAN). Synthetically generated MRI increased the training accuracy from 78.9% to 93% and test accuracy from 69.4% to 79.5% in this study.Introduction
Mutation in neurofibromatosis type 2 (NF2) gene is one of the biomarkers of tumorigenesis in meningiomas that play an important role in predicting prognosis 1,2. Noninvasive detection of NF2 mutation prior to surgery would help with better treatment planning of meningiomas3. Although deep learning methods have shown some success for a large span of medical image classification problems, there are still some obstacles and limitations that prevent a wider usage in clinical practice. One of the main limitations of deep learning applications is the amount of annotated data that is necessary for training deep networks. Besides the conventional data augmentation methods, such as reflection, rotation and translation, generative adversarial networks (GAN) have been commonly employed4. The aims of this study are to generate synthetic T1-weighted contrast-enhanced (T1C) MRI of meningiomas to help with the data insufficiency problem, and to assess the effects of the synthetic data on the performance of deep learning methods for the classification of NF2 copy number loss status in meningiomas.Methods
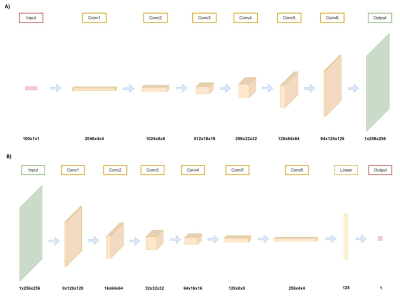
Eighty-three meningioma patients (53F/30M, mean age = 51.13 ± 14.11 years, 39 NF2 loss, 44 NF2 intact) were included in this study. The patients were scanned on a 3T Siemens MR scanner (Erlangen, Germany) using a 32-channel head coil prior to surgery. The brain tumor protocol included pre- and post-contrast (gadolinium DTPA) T1-weighted TSE (TR=500 ms, TE=10 ms) for preoperative patients. NF2-copy number loss was determined by digital droplet PCR using pre-validated Taq-man probes. The tumor regions were delineated and cropped on T1C MRI. The resulting 2D images were padded and reshaped into 256x256 frames. For the synthetic image generation, a deep convolutional generative adversarial network (DCGAN)5 was implemented in PyTorch 1.11 (Meta Platforms Inc., Menlo Park, CA) framework (Figure 1). T1C MRI of NF2 loss and NF2 no-loss groups were separated, and synthetic MRI were generated for each group using the DCGAN model. For the classification task, a pre-trained ResNet506 model was used to initialize the weights. Except for the last convolutional block, the weights of all the layers were fixed, and a fully connected layer with 512 units was added to the end of the network. Hyperparameters of the model were optimized using Weights & Biases: Sweep module7. Binary cross-entropy was used as the loss function and Adam optimizer was used for the optimization at best scenario. The classification task was performed twice by training with only the original T1C MRI followed by using both the original and augmented data, and the accuracy, sensitivity (recall), specificity, and precision metrics were compared.Results
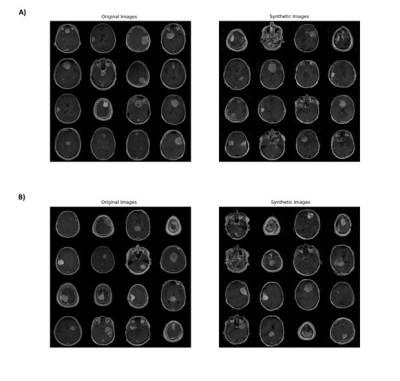
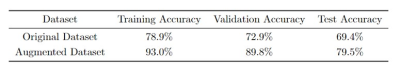
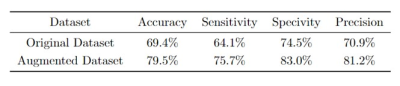
For the classification model trained with the original dataset, meningiomas with NF2 loss were classified with 78.9% accuracy for the training, 72.9% for the validation and 69.4% for the test datasets (sensitivity=64.1%, specificity=74.5%, precision=70.9% for the test data) (Tables 1-2). On the other hand, using augmented data (Figure 2) increased the classification accuracies to 93% for the training, 89.8% for the validation and 79.5% for the test datasets (sensitivity=75.7%, specificity=83%, precision=81.2% for the test data) (Tables 1-2).Conclusion and Discussion
The results of this study indicated that the proposed DCGAN model could produce high-quality T1C MRI of meningiomas. The resulting augmented images were used to improve the classification accuracy of NF2 copy number loss status in meningiomas. Future studies will test GAN models for improving the prediction accuracy for other genetic markers in meningiomas.Acknowledgements
This study was supported by the Scientific and Technological Research Council of Turkey (TUBITAK) grant 119S520.References
1. Al-Rashed M, Foshay K, Abedalthagafi M. Recent Advances in Meningioma Immunogenetics. Front Oncol 2019;9:1472.
2. Lee S, Karas PJ, Hadley CC, Bayley V JC, Khan AB, Jalali A, et al. The Role of Merlin/NF2 Loss in Meningioma Biology. Cancers (Basel) 2019;11(11):1633.
3. Aboukais R, Zairi F, Baroncini M, Bonne NX, Schapira S, Vincent C, et al. Intracranial meningiomas and neurofibromatosis type 2. Acta Neurochir (Wien) 2013;155(6):997-1001.
4. Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets. Advances in neural information processing systems 2014;27.
5. Radford A, Metz L, Chintala S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv preprint arXiv:151106434 2015.
6. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition. 770-8
7. Biewald L. Experiment tracking with weights and biases. Software available from wandb com 2020;2:233.
Figures