5032
Deep learning-based synthesis of TSPO PET from T1-weighted MRI images only
Matteo Ferrante1, Marianna Inglese2, Ludovica Brusaferri3, Marco L Loggia3, and Nicola Toschi4,5
1Biomedicine and prevention, University of Rome Tor Vergata, Roma, Italy, 2Biomedicine and prevention, University of Rome Tor Vergata, Rome, Italy, 3Martinos Center For Biomedical Imaging, MGH and Harvard Medical School (USA), Boston, MA, United States, 4BioMedicine and prevention, University of Rome Tor Vergata, Rome, Italy, 5Department of Radiology,, Athinoula A. Martinos Center for Biomedical Imaging and Harvard Medical school, Boston, MA, USA, Boston, MA, United States
1Biomedicine and prevention, University of Rome Tor Vergata, Roma, Italy, 2Biomedicine and prevention, University of Rome Tor Vergata, Rome, Italy, 3Martinos Center For Biomedical Imaging, MGH and Harvard Medical School (USA), Boston, MA, United States, 4BioMedicine and prevention, University of Rome Tor Vergata, Rome, Italy, 5Department of Radiology,, Athinoula A. Martinos Center for Biomedical Imaging and Harvard Medical school, Boston, MA, USA, Boston, MA, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Neuroinflammation, PET, image synthesis
Chronic pain-related biomarkers can be found using a specific binding radiotracer called [11C]PBR28 able to target the translocator protein (TSPO), whose expression is increased in activated glia and can be considered as a biomarker for neuroinflammation. One of the main drawbacks of PET imaging is radiation exposure, for which we attempted to develop a deep learning model able to synthesize PET images of the brain from T1w MRI only. Our model produces synthetic TSPO-PET images from T1W MRI which are statistically indistinguishable from the original PET images both on a voxel-wise and on a ROI-wise level.Introduction
Chronic pain is a common condition with unknown etiology, and difficult to assess through exams or medical imaging. In animals, there is evidence that persistent pain is related to an inflammation of cells in the brain like microglia and astrocytes. In humans, there is recent evidence that neuroinflammation can be related to chronic pain (Loggia et al., 2015) for a variety of pain conditions including lower back pain, amyotrophic lateral sclerosis, knee osteoarthosis and many more. Research in this field revolves around devising the most sensitive and specific strategy to detect and measure inflammation. Neuroinflammation of glial cells can be detected using translocator protein (TSPO) radiotracer binding ([11C]PBR-28). Recent findings have involved the thalamus in a reproducible neuroinflammation signature in chronic pain. However, PET imaging is expensive, requires the use of ionizing radiation and being close to cyclotron. Structural (i.e. T1-w) images may encode information relevant to neuroinflammation because the latter also involves some morphological/structural changes in the glial cells. Moreover, a model able to generate these images could pave the way to large retrospective studies on structural images. In this work we aim to recover TSPO-PET images using T1-w MR imaging exclusively, hence foregoing the use of ionizing radiation. On the basis of previous work for other radiotracers image synthesis(Sikka et al., 2021; Zhang et al., 2022), We propose a modality conversion, deep learning model based on U-Net which is able to generate synthetic brain PET images starting from a subject’s T1-w scan only.Data
The dataset employed in this study included 204 patients who underwent 3T MR-PET ([11C]PBR28 radiotracer) scans, including 28 healthy controls (HC) as well as 89 knee-osteoarthritis (KOA) and 87 chronic lower back pain patients (CLB). Prior to PET-MR imaging, all subjects were injected with a dose in the range 9-15 mCi. T1-weighted and PET images were coregistered, skull-stripped, and normalized using FSL and python. Brain maks were also retained and used to mask the PET and T1 images prior to model training. The data was randomly divided in training and test set with an 80-20 proportion, and the model was trained to map one modality to the other. The training inputs were subject-wise pairs of normalized T1 and PET SUV images, and the output on the unseen test set were synthetic PET images.Model
Our model was a 3D U-net (Weng & Zhu, 2021)-based architecture which exploits depthwise separable convolutions and attention mechanisms. The architecture is based on 4 layers of downsampling based on depthwise convolutions with 32 channels each. These are used to minimize the number of parameters to deal with 3D images without overfitting or generating prohibitive memory footprints. In the last layer, and attention mechanism is applied in order to couple the local inductive bias of a convolutional neural network with a global latent attention to take into account possible long-range dependencies. The model is trained with a balanced loss between binary cross-entropy and mean squared error for 50 epochs using the Adam optimizer.Results
We evaluated a) statistical differences between original and synthetic images in the test set, 2) to what extent group-wise differences found in the original PET data were preserved in the synthetic data. The first point was evaluated through nonparametric voxel- and ROI-based analysis, while the second was evaluated in a ROI-based manner on the thalamus defined in the native space, which has recently been shown to exhibit increased neuroinflammation in chronic pain. Voxel-wise analysis using the randomize permutation (N=10000) tool in FSL (paired t-test) showed no significant differences between original and synthetic PET data, which was confirmed by thalamic ROI-based analysis across the whole brain. Also, the average signal in the thalamus statistically different in the following pairwise comparisons (Mann Whitney U-test): HC vs CLB (p value original 0.95, p value synthetic 0.95), HC vs KOA (p value original 0.21, p value synthetic 0.03) and CLB and KOA (p value original 0.07, p value synthetic 0.10). when using both original and synthetic images. Interestingly, the median signal value in the thalamus was significantly different between KOA and HC only in the synthetic images.Conclusions
Our model is able to generate synthetic PET images from structural T1w images with the same spatial signal distribution of signal as the original images. Qualitatively, the synthetic images appeared smoother than the original ones, and hence extremely similar to the original images after a smoothing gaussian kernel is applied (see Figure). We hypothesize that model learns predominantly signal conversion from one modality to another from the statistics of the training set, discarding noise components and hence acting as an inherent denoising operator. This can potentially enhance the signal-to-noise ratio (SNR), possibly enhancing slight group-differences which reached statistical significance when using synthetic PET only. This would qualify our model as a tool to also increase sensitivity in the synthetic modality which is generated from the T1 image. Our synthetic PET images are statistically indistinguishable from the original data and preserve the original sensitivity to pathophysiological changes. This means that there is no systematic shift in the synthetic data generated and suggests that our model has the potential to replace radioisotopic imaging with MR data only.Acknowledgements
Part of this work is supported by the EXPERIENCE project (European Union’s Horizon 2020 research and innovation program under grant agreement No. 101017727)Matteo Ferrante is a Ph.D. student enrolled in the National PhD in Artificial Intelligence, XXXVII cycle, course on Health and life sciences, organized by Università Campus Bio-Medico di Roma.References
Loggia, M. L., Chonde, D. B., Akeju, O., Arabasz, G., Catana, C., Edwards, R. R., Hill, E., Hsu, S., Izquierdo-Garcia, D., Ji, R. R., Riley, M., Wasan, A. D., Zurcher, N. R., Albrecht, D. S., Vangel, M. G., Rosen, B. R., Napadow, V., & Hooker, J. M. (2015). Evidence for brain glial activation in chronic pain patients. Brain, 138(3), 604–615. https://doi.org/10.1093/brain/awu377 Sikka, A., Skand, Virk, J. S., & Bathula, D. R. (2021). MRI to PET Cross-Modality Translation using Globally and Locally Aware GAN (GLA-GAN) for Multi-Modal Diagnosis of Alzheimer’s Disease. XX(Xx), 1–12. http://arxiv.org/abs/2108.02160 Weng, W., & Zhu, X. (2021). INet: Convolutional Networks for Biomedical Image Segmentation. IEEE Access, 9, 16591–16603. https://doi.org/10.1109/ACCESS.2021.3053408 Zhang, J., He, X., Qing, L., Gao, F., & Wang, B. (2022). BPGAN: Brain PET synthesis from MRI using generative adversarial network for multi-modal Alzheimer’s disease diagnosis. Computer Methods and Programs in Biomedicine, 217, 106676. https://doi.org/10.1016/j.cmpb.2022.106676Figures
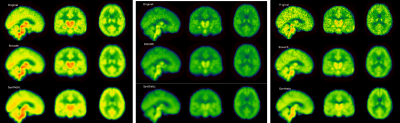
Results. From left to right: Average results for
Chronic Lower Back Pain patients, Healthy controls and Knee Osteoarthrosis
patients in the test set. Original: Normalized PET image. Smooth: normalized
image after smoothing. Synthetic: is the corresponding image generated from
T1-w MRI only. The spatial distribution of signal is clearly consistent across
different patient populations.
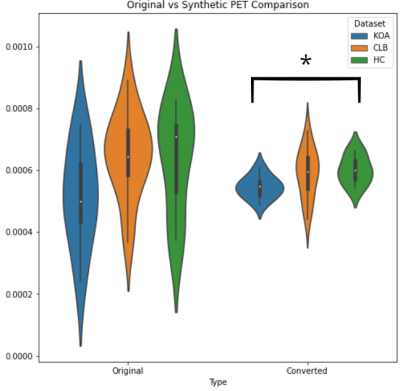
Differences
between original and synthetic PET among different patient groups in the thalamic
region. Converted (Synthetic) images shown differences between KOA and HC, probably
due to noise and variance reduction done by the model as an
emergent property.
DOI: https://doi.org/10.58530/2023/5032