5013
Novel light-sheet scattering microscopy for voxel-wise validation of 3D-fibre orientations in murine white matter1Department of Applied Mathematics and Computer Science, Technical University of Denmark, Kongens Lyngby, Denmark, 2Danish Research Centre for Magnetic Resonance (DRCMR), Centre for Functional and Diagnostic Imaging and Research, Copenhagen University Hospital, Amager & Hvidovre, Copenhagen, Denmark, 3Gubra, Hørsholm, Denmark, 4Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute for Science and Technology (BIST), Barcelona, Spain
Synopsis
Keywords: Validation, Tractography & Fibre Modelling, 3D-Fibre orientation
This work develops a novel 3D validation technique that reveals the white matter orientation with micron resolution in cleared murine brains. Light-sheet elastic scattering microscopy (LSSM) can be used to exploit the scattering signal exhibited by white matter fibres, which is dependent on their orientation. The rotation and imaging of the sample yield a scattering profile, which corresponds to that of infinitely long cylinders. Our work combines two orthogonal acquisitions to voxel-wise reconstruct 3D-fibre orientations in large brain volumes. LSSM could be the definitive validation method for diffusion MRI orientation methods.Introduction
Diffusion MRI tractography is the only imaging technique that allows for non-invasive mapping of the brain network. However, the resolution of dMRI is insufficient to grasp the full complexity of the brain. Hence, downstream applications like tractography and structural connectivity analysis are subject to multiple uncertainties1,2.Alternative fibre orientation contrasts are crucial to validate the findings of dMRI and bring tractography methods toward the next generation. Structure tensor analysis3 and other recent microscopy systems4,5,6 enable the estimation of fibre orientations at a sub-MRI voxel level. However, previous techniques require slicing the sample, limiting the field of view. Moreover, the analysis is usually limited to the 2D plane and no technique reveals the full volume 3D-fibre orientation natively. We propose 3D-light-sheet scattering microscopy (LSSM) to map fibre orientation in micron resolution of cleared-tissue samples. This approach is label-free, does not require histological sectioning and can unravel the actual 3D-fibre orientation in large brain volumes.
Methods and Materials
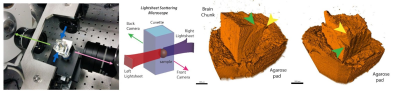
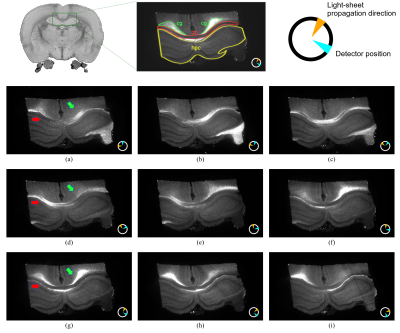
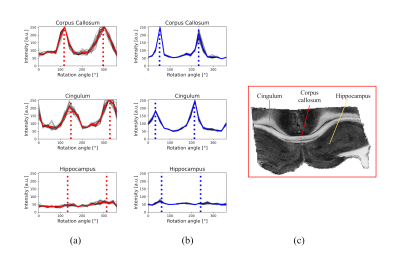
In light-sheet microscopy (LSM) a plane of interest is illuminated with a thin sheet of laser light, while a detection objective is placed orthogonally to it. The sample is optically cleared (made chemically transparent to light) using the iDISCO+ protocol7 and a volumetric image can be formed by moving the cuvette holding the sample along the axis of detection. Our microscope (MacroSPIM) setup counts with two opposed illumination arms, and two opposed detection objectives (Fig. 1).LSM has been typically used for fluorescence imaging (LSFM). However, we found that imaging the laser lightsheet signal scattered by brain tissues, consistently with lightsheet elastic scattering8 or lightsheet scattering microscopy9, reveals a fibre-dependent optical signal congruent with neural pathways, and dependent on anisotropic tissue orientation (Fig. 2). Hence, we address the fibre angular dependence of the signal by rotating the sample 18 times to cover in-plane the 360$$$^\circ$$$ angular range, and we obtain a scatter profile with two peaks per fibre direction. A shift and amplitude modulation of the peaks rely on the 3D sample orientation relative to incident light. This behaviour adheres ostensibly to the solution of Maxwell’s equations for the scattering of light by an infinite cylinder10,11. When the light hits a fibre (cylinder), it scatters into a conical surface, whose aperture depends on the angle between the fibre and the incident light. Thus, the model for a single fibre (axon) predicts a peak signal intensity where the detectable scatter wavefronts are directed toward the detector. The intensity modulation reflects the tilting (out-of-plane directionality) of the fibres, showing larger values for fibres orthogonal to the rotation axis.
This work assesses the feasibility of this approach on a block of murine’s brain tissue including the corpus callosum and the cingulum, imaged at optically moderate resolution (5x3x3 µm3), yet very high compared to typical dMRI resolution. In total, every acquisition comprises 4x18 3D image volumes, each of 10.5 GB.
Results
The comparison between simulated and experimental scattering profiles reveals a good matching between the peaks' position and the in-plane directionality, while the amplitude of the peaks is not a solid indicator of the tilting as expected. For example, corpus callosum and cingulum tracts show similar amplitude modulation despite having well-described in-plane and out-of-plane orientations (Fig. 3). Since LSSM enables us to mount the sample in any orientation, we manually rotated the sample 90 degrees and repeated the acquisition. As a result of 3D registration and integration of the 2D information from the two orthogonal-like acquisitions (rotations planes), we can estimate the tilting of the fibre independently of the amplitude modulation. Now, we can realize a voxel-wise 3D-fiber orientation reconstruction (Fig. 4). The estimated orientations, illustrated here with 1 pair of detector/lightsheet, are consistent across the 4 possible pairs (front/back detector, right/left lightsheet), indicating the stability of the method. The scattering profiles in the grey matter do not show prominent peaks.Discussion
We show a novel imaging workflow with LSSM for cleared-brain tissues that enables reliable high-resolution whole-brain fibre orientation estimation in 3D like dMRI but with optical resolution. Theoretically, according to the scattering of light by infinite cylinders, we should be able to unravel the 3D fibre orientation with a single-rotation acquisition. In practice, deciphering the relationship between the amplitude modulation and the tilting of the fibre while imaging a sample in a single position is challenging. The tilting angle to obtain the actual 3D orientation can be easily solved with an additional rotation of the sample. This is not possible with other histological slice-based optical techniques like PLI and SCI4,6. Future research should explore additional opportunities for single-rotation acquisitions. Uniquely, the LSSM dataset could be aligned to diffusion MRI of the same brain by first collecting MRI and then LSSM12.Conclusion
We show that LSSM of intact cleared-tissue samples is sensitive to 3D fibre orientation similarly to diffusion-based MRI, albeit with a different physical foundation. Our preliminary results confirm the feasibility and solidity of the suggested technique, here applied at intermediate optical magnification (>10x), only limited by the microscope and the amount of data to be handled. This approach could play a key role in the validation of fibre orientation and tractography algorithms and provide new detailed insights into the brain network.Acknowledgements
The work is supported by a research grant (17493) from VILLUM FONDEN and by a research grant (R370-2021-1010) from Lundbeckfonden (PI: Tim Dyrby).References
1. Klaus H. Maier-Hein et al. “The challenge of mapping the human connectome based on diffusion tractography”. In: Nature Communications 2017 8:1 8.1 (Nov. 2017), pp. 1–13. ISSN: 2041-1723. DOI: 10.1038/s41467- 017- 01285- x. URL: https://www.nature.com/articles/ s41467-017-01285-x.
2. Kurt G. Schilling et al. “Tractography dissection variability: What happens when 42 groups dissect 14 white matter bundles on the same dataset?” In: NeuroImage 243 (Nov. 2021), p. 118502. ISSN: 1053-8119. DOI: 10.1016/J.NEUROIMAGE.2021.118502.
3. Kurt G. Schilling et al. “Histological validation of diffusion MRI fiber orientation distributions and dispersion”. In: NeuroImage 165 (Jan. 2018), pp. 200–221. ISSN: 1053-8119. DOI: 10.1016/J. NEUROIMAGE.2017.10.046.
4. Markus Axer et al. “High-resolution fiber tract reconstruction in the human brain by means of three-dimensional polarized light imaging”. In: Frontiers in Neuroinformatics 5 (Dec. 2011), p. 34. ISSN: 16625196. DOI: 10.3389/FNINF.2011.00034/BIBTEX.
5. Hui Wang et al. “Cross-validation of serial optical coherence scanning and diffusion tensor imaging: A study on neural fiber maps in human medulla oblongata”. In: NeuroImage 100 (Oct. 2014), pp. 395–404. ISSN: 1053-8119. DOI: 10.1016/J.NEUROIMAGE.2014.06.032.
6. Miriam Menzel et al. “Scattered Light Imaging: Resolving the substructure of nerve fiber crossings in whole brain sections with micrometer resolution”. In: NeuroImage 233 (June 2021). ISSN: 10959572. DOI: 10.1016/J.NEUROIMAGE.2021.117952.
7. Nicolas Renier et al. “Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes”. In: Cell 165.7 (June 2016), pp. 1789–1802. ISSN: 0092-8674. DOI: 10.1016/J. CELL.2016.05.007.
8. Diego Di Battista et al. “Enhanced Light Sheet Elastic Scattering Microscopy by Using a Supercontinuum Laser”. In: Methods Protoc. 2, 57 (2019) DOI: 10.3390/mps2030057
9. Xiangda Zhou et al. “Light-Sheet Scattering Microscopy to Visualize Long-Term Interactions Between Cells and Extracellular Matrix”. In: Front.Immunol.13:828634. (2022) DOI: 10.3389/fimmu.2022.828634
10. Hashim A. Yousif and Edward Boutros. “A FORTRAN code for the scattering of EM plane waves by an infinitely long cylinder at oblique incidence”. In: Computer Physics Communications 69.2-3 (Mar. 1992), pp. 406–414. ISSN: 0010-4655. DOI: 10.1016/0010-4655(92)90178-2.
11. Jan Schäfer, et al. "Calculation of the near fields for the scattering of electromagnetic waves by multiple infinite cylinders at perpendicular incidence". In: Journal of Quantitative Spectroscopy and Radiative Transfer, 113.16 (Nov. 2012), pp. 2113-2123. ISSN: 0022-4073. DOI: 10.1016/j.jqsrt.2012.05.019.
12. Johanna Perens et al. “Multimodal 3D mouse brain atlas framework with skull-derived coordinate system”. In: Research Square (Jul. 2022) [Preprint, VERSION 1]. DOI: 10.21203/rs.3.rs-1832101/v1. URL: https://doi.org/ 10.21203/rs.3.rs-1832101/v1.
Figures