4985
Recovering image orientation for unconventional acquisition plane with deep learning algorithm: cardiac magnetic resonance case.
Habib Rebbah1 and Timothé Boutelier1
1Research & Innovation, Olea Medical, La Ciotat, France
1Research & Innovation, Olea Medical, La Ciotat, France
Synopsis
Keywords: Myocardium, Segmentation
Deducing the orientation main view of an image from the DICOM information could be complicated especially for unconventional image plane as for cardiac MR. We explore here the feasibility of deducing the orientation of the image based on CNN approach.
Introduction
For many medical image processing algorithms, it could be advantageous to merge the images to process into the same orientation, like for instance putting the pre-frontal cortex at the top of the image . Such a correction is often met for brain images processing, while the acquisitions are done based on conventional planes (sagittal, coronal, and transverse). Indeed, from the image orientation and position information stored in the DICOM fields, one could adapt the orientation of images to match the desired one. However, for many applications, these transformations are not trivial due to unconventional acquisition planes. Furthermore, it could be recommended basing the correction only on image information since some databases do not include DICOM’s header details. Here, we treat the cardiac magnetic resonance (CMR) case and propose to explore the feasibility of deducing the orientation of the image based on deep learning approach.Methods
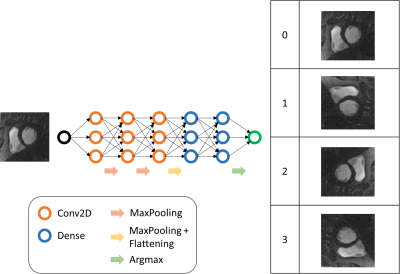
The algorithm is a convolutionnal neural network (CNN) and takes as input a 2D CMR image and returns a label corresponding to its main orientation. We allocated the orientations in 4 classes separate by π/2 rotations. The figure 1 illustrates the approach.We trained the neural network with images from CINE, perfusion, MOLLI pre-injected, MOLLI post-injected and T2 prepared (T2p) sequences to obtain 5 algorithms for each sequence. Each dataset was tacking from the HIBISCUS-STEMI database (NCT03070496) and incorporates a homogeneous repartition of slice positions from the base to the apex of the heart. All the datasets were constituted of 0 and 1 classes of orientations. Therefore, we added a random π/2 rotations to obtain a homogenous distribution of classes orientation.
Since all the sequences processed are time series sequences, the input 2D image was the first image for CINE and T2p, the last one for MOLLIs, and the image with highest contrast (evaluated as the standard deviation -sd- of the image) for perfusion. A ratio of 80/20% for training/evaluation subdivision of datasets was used for all the fitting processes. The table 1 resumes the methodology.
The networks were built using Tensorflow framework and all the developments were done in Python.
Results and discussion
The table 2 gather all the results in term of accuracy for the validation part of each dataset. The lowest accuracy value observed is 0.997 while the highest is 0.999.These results allow to use these algorithms as a pre-processing step to merge the input images into the same desired view, reducing then the variability. Such an automatic processing is required to avoid additional manual requested steps, potentially source of bias. In our case this harmonization allows us to train segmentation algorithms that do not involve some data augmentation steps, hence reducing the uncertainty.
Conclusion
The CNN approach successfully identified the image orientation in the CMR quantification case.Acknowledgements
This work was supported by the RHU MARVELOUS (ANR-16-RHUS-0009). The authors particularly thank Pierre Croisille, Magalie Viallon, Nathan Mewton, Charles De Bourguignon, and Lorena Petrusca for data acquisition and management.References
No reference found.
DOI: https://doi.org/10.58530/2023/4985