4981
Accelerated, Free-Breathing 3D Cardiac T1ρ Mapping Pulse Sequence with XD-GRASP Reconstruction1Radiology, Northwestern University, Feinberg School of Medicine, Chicago, IL, United States, 2Cardiology, Lurie Children’s Hospital, Chicago, IL, United States, 3Cardiology, Northwestern University, Feinberg School of Medicine, Chicago, IL, United States, 4Electrical and Computer Engineering, Northwestern University, Evanston, IL, United States, 5Radiology, Lurie Children’s Hospital, Chicago, IL, United States, 6Radiology, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, United States
Synopsis
Keywords: Myocardium, Quantitative Imaging, Fibrosis
T1ρ mapping is an emerging non-contrast pulse sequence for measuring cardiac fibrosis. Unfortunately, current T1ρ techniques suffer from lack of coverage, poor spatial resolution, and long scan time. Thus, we developed an accelerated, free-breathing 3D cardiac T1ρ mapping pulse sequence using XD-GRASP reconstruction including both respiratory and contrast dimensions; additionally, view sharing and KWIC filtering were incorporated to improve spatial resolution. This sequence was tested in 8 patients undergoing clinically indicated cardiac MRI, resulting in robust image quality and T1ρ values that are in agreement with literature.Introduction
T1ρ mapping is an emerging non-contrast pulse sequence that can be used to detect both focal1-3 and diffuse4, 5 myocardial fibrosis. Despite its enormous potential for assessment of myocardial fibrosis in pediatric patients and adult patients with renal conditions, cardiac T1ρ mapping suffers from several technical limitations including poor spatial resolution and long scan time (up to 18 min)6, 7. Therefore, in this study, we sought to address these limitations of 3D cardiac T1ρ mapping by developing a stack-of-stars k-space sampling pattern and XD-GRASP8 reconstruction. Our new pulse sequence is initially evaluated in control patients with no fibrosis.Methods
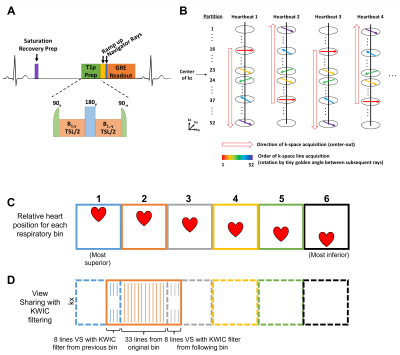
Human Subjects: The study included 8 patients (mean age 70 ± 14 years; 4 females) who underwent clinically scheduled cardiac MRI at 1.5 T (MAGNETOM Avanto, Siemens); clinical MRI protocol included native T1 imaging, extra cellular volume fraction (ECV) mapping, 2D late gadolinium enhancement (LGE), and when clinically indicated, T2 mapping. We added the proposed 3D T1ρ mapping prior to contrast agent administration.Pulse Sequence & Image Reconstruction: We modified a previously described 3D LGE pulse sequence9 using a stack-of-stars k-space sampling pattern and XD-GRASP reconstruction8; in the modified sequence, we added a B1-insensitive saturation pulse10 to reset the longitudinal magnetization for each heartbeat prior to an updated adiabatic half passage (AHP) T1ρ preparation pulse (90x(AHP) – B1y – 180y – B1-y – 90-x(AHP)) with B1 = 500 Hz6 (Figure 1A) and 3 T1ρ-weighting spin-lock times (TSL = 0, 35, 50 ms). Data were collected alternating center-out to fill 52 partitions of the 3D volume (Figure 1B). Prior to each single-shot GRE readout (flip angle = 15°), two rays were sampled for self‐navigation of respiratory motion11. 3D T1ρ mapping data were imaged in a coronal view with field-of-view of 288-316 mm x 288-316mm x 114 mm and spatial resolution of 1.5-2.2 mm x 1.5-2.2 mm x 2.2 mm. In total 600 heartbeats (200 heartbeats for each TSL 3D image) were scanned. To improve spatial resolution, view sharing (VS)12 of 8 radial spokes from adjacent respiratory bins and k-space weighted image contrast (KWIC)13 filtering was used (Figure 1D). The 3D T1ρ data were reconstructed using the XD-GRASP framework8 with both respiratory and TSL contrast as extra dimensions.
Image Processing: T1ρ maps were generated from the images by fitting a two parameter signal model given by S=S0e(−TSL/T1ρ)14 where S0 is the initial magnetization after the saturation pulse and TSL is the spin-lock pulse duration, using MATLAB (MathWorks, Natick, MA). Myocardial contours were manually drawn for each slice within the 3D volume and averaged over all slices using a MATLAB program developed in our lab. American Heart Association (AHA) 16-segment bullseye model15 of T1ρ was calculated by reformatting the 3D volume was in RadiAnt Dicom Viewer (Medixant, Pozan, Poland).
Results
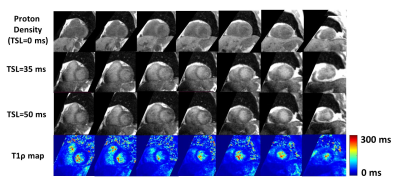
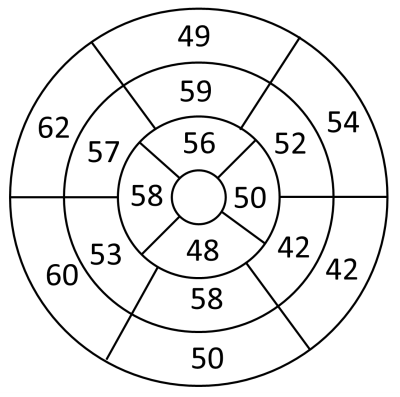
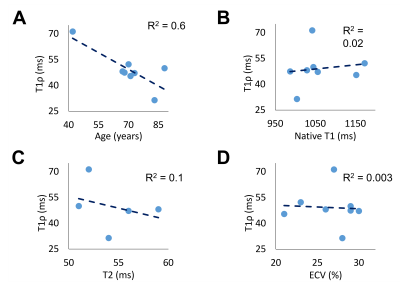
3D T1ρ mapping took an average of 12 ± 2 minutes to complete a 3D volume (Figure 2). Mean metrics were: T1ρ = 49 ± 11 ms, native T1 = 1061 ± 67 ms, ECV = 27 ± 3%, T2 = 54 ± 3 ms which corresponds to normal values6, 7, 16-18. In the patient group, none had focal scar on LGE examination. The AHA 16 segment model demonstrated higher T1ρ values in the septum compared to the lateral side (Figure 4). Mean T1ρ was plotted against patient age, native T1, T2, and ECV which resulted in no correlation with T2 and ECV values, but there was a slight trend with T1ρ increasing with native T1 and T1ρ decreasing with age (Figure 5).Discussion
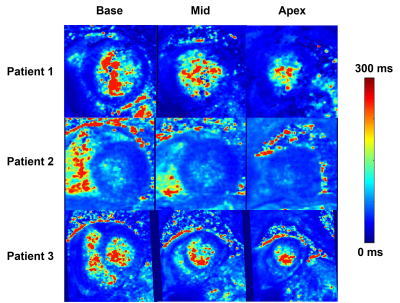
This study demonstrates that our proposed accelerated, high-resolution 3D T1ρ using VS and KWIC produced excellent image quality in patients at 1.5T. We demonstrate that T1ρ maps were achieved in each slice of the acquired 3D volume (Figure 2) and that this data quality is reproducible across different patients (Figure 3). T1ρ maps resulted in average T1ρ values that are in the same range as previously published values6, 7. While trends were seen with T1ρ and native T1 and age, a larger patient group is needed to derive statistical significance. The AHA 16 segment model demonstrated higher T1ρ values in the septum compared to the lateral side, but this could be due to potential partial volume averaging from reformatting. Since the ECV of our cohort was in the normal range and none of the patients had focal scar, we did not detect fibrosis. Thus, this cohort could serve as controls for future studies including patients with fibrosis. A limitation of this study is that we did not compare our T1ρ against reference T1ρ, which does not exist. Future studies include scanning patients with fibrosis, including a 2D T1ρ pulse sequence as reference, further optimizing 3D T1ρ pulse sequence and reconstruction parameters, and incorporation of deep learning methods for automated quantification of 3D T1ρ maps.Conclusion
3D T1ρ mapping using stack-of-stars k-space sampling and XD-GRASP reconstruction with view sharing and KWIC filtering achieves robust image quality and T1ρ values that agree with literature and could be used for further studies for non-contrast fibrosis imaging.Acknowledgements
This work was supported in part by the following grants: National Institutes of Health (1K99HL161469, R01HL116895, R21AG055954, R01HL151079, R21EB030806A1) and American Heart Association (19IPLOI34760317, 949899)References
1. Muthupillai, R.; Flamm, S. D.; Wilson, J. M.; Pettigrew, R. I.; Dixon, W. T., Acute myocardial infarction: tissue characterization with T1rho-weighted MR imaging--initial experience. Radiology 2004, 232 (2), 606-10.
2. Witschey, W. R.; Zsido, G. A.; Koomalsingh, K.; Kondo, N.; Minakawa, M.; Shuto, T.; McGarvey, J. R.; Levack, M. M.; Contijoch, F.; Pilla, J. J.; Gorman, J. H., 3rd; Gorman, R. C., In vivo chronic myocardial infarction characterization by spin locked cardiovascular magnetic resonance. J Cardiovasc Magn Reson 2012, 14, 37.
3. van Oorschot, J. W.; El Aidi, H.; Jansen of Lorkeers, S. J.; Gho, J. M.; Froeling, M.; Visser, F.; Chamuleau, S. A.; Doevendans, P. A.; Luijten, P. R.; Leiner, T.; Zwanenburg, J. J., Endogenous assessment of chronic myocardial infarction with T(1rho)-mapping in patients. J Cardiovasc Magn Reson 2014, 16, 104.
4. van Oorschot, J. W.; Guclu, F.; de Jong, S.; Chamuleau, S. A.; Luijten, P. R.; Leiner, T.; Zwanenburg, J. J., Endogenous assessment of diffuse myocardial fibrosis in patients with T1rho -mapping. J Magn Reson Imaging 2017, 45 (1), 132-138.
5. Zhang, Y.; Zeng, W.; Chen, W.; Chen, Y.; Zhu, T.; Sun, J.; Liang, Z.; Cheng, W.; Wang, L.; Wu, B.; Gong, L.; Ferrari, V. A.; Zheng, J.; Gao, F., MR extracellular volume mapping and non-contrast T1rho mapping allow early detection of myocardial fibrosis in diabetic monkeys. Eur Radiol 2019, 29 (6), 3006-3016.
6. Kamesh Iyer, S.; Moon, B.; Hwuang, E.; Han, Y.; Solomon, M.; Litt, H.; Witschey, W. R., Accelerated free-breathing 3D T1rho cardiovascular magnetic resonance using multicoil compressed sensing. J Cardiovasc Magn Reson 2019, 21 (1),5.
7. Qi, H.; Bustin, A.; Kuestner, T.; Hajhosseiny, R.; Cruz, G.; Kunze, K.; Neji, R.; Botnar, R. M.; Prieto, C., Respiratory motion-compensated high-resolution 3D whole-heart T1rho mapping. J Cardiovasc Magn Reson 2020, 22 (1), 12.
8. Gunasekaran S, H. H. H.-V., R Passman, D Lee, D Kim In Self-Navigated, Free-Breathing 3D Left Atrial Late Gadolinium Enhancement MRI with Stack-of-Stars k-space Sampling and GRASP Reconstruction: A Preliminary Study for Quantification of Atrial Fibrosis, ISMRM, Montreal, QC, Canada, Montreal, QC, Canada, 2019.
9. Gunasekaran, S.; Haji-Valizadeh, H.; Lee, D. C.; Avery, R. J.; Wilson, B. D.; Ibrahim, M.; Markl, M.; Passman, R. S.; Kholmovski, E. G.; Kim, D., Accelerated 3D Left Atrial Late Gadolinium Enhancement in Patients with Atrial Fibrillation at 1.5 T: Technical Development. Radiol Cardiothorac Imaging 2020, 2 (5), e200134.
10. Kim, D.; Oesingmann, N.; McGorty, K., Hybrid adiabatic-rectangular pulse train for effective saturation of magnetization within the whole heart at 3 T. Magn Reson Med 2009, 62 (6), 1368-78.
11. Haji-Valizadeh, H.; Collins, J. D.; Aouad, P. J.; Serhal, A. M.; Lindley, M. D.; Pang, J.; Naresh, N. K.; Carr, J. C.; Kim, D., Accelerated, free-breathing, noncontrast, electrocardiograph-triggered, thoracic MR angiography with stack-of-stars k-space sampling and GRASP reconstruction. Magn Reson Med 2019, 81 (1), 524-532.
12. Foo, T. K.; Bernstein, M. A.; Aisen, A. M.; Hernandez, R. J.; Collick, B. D.; Bernstein, T., Improved ejection fraction and flow velocity estimates with use of view sharing and uniform repetition time excitation with fast cardiac techniques. Radiology 1995, 195 (2), 471-8.
13. Song, H. K.; Dougherty, L., k-space weighted image contrast (KWIC) for contrast manipulation in projection reconstruction MRI. Magn Reson Med 2000, 44 (6), 825-32.
14. Witschey, W. R.; Borthakur, A.; Elliott, M. A.; Fenty, M.; Sochor, M. A.; Wang, C.; Reddy, R., T1rho-prepared balanced gradient echo for rapid 3D T1rho MRI. J Magn Reson Imaging 2008, 28 (3), 744-54.
15. Cerqueira, M. D.; Weissman, N. J.; Dilsizian, V.; Jacobs, A. K.; Kaul, S.; Laskey, W. K.; Pennell, D. J.; Rumberger, J. A.; Ryan, T.; Verani, M. S.; American Heart Association Writing Group on Myocardial, S.; Registration for Cardiac, I., Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. A statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Int J Cardiovasc Imaging 2002, 18 (1), 539-42.
16. Reiter, U.; Reiter, G.; Dorr, K.; Greiser, A.; Maderthaner, R.; Fuchsjager, M., Normal diastolic and systolic myocardial T1 values at 1.5-T MR imaging: correlations and blood normalization. Radiology 2014, 271 (2), 365-72.
17. Haaf, P.; Garg, P.; Messroghli, D. R.; Broadbent, D. A.; Greenwood, J. P.; Plein, S., Cardiac T1 Mapping and Extracellular Volume (ECV) in clinical practice: a comprehensive review. J Cardiovasc Magn Reson 2016, 18 (1), 89.
18. Meloni, A.; Nicola, M.; Positano, V.; D'Angelo, G.; Barison, A.; Todiere, G.; Grigoratos, C.; Keilberg, P.; Pistoia, L.; Gargani, L.; Ripoli, A.; Pepe, A., Myocardial T2 values at 1.5 T by a segmental approach with healthy aging and gender. European Radiology 2022, 32 (5), 2962-2975.
Figures