4975
2D U-Net based myocardium segmentation for quantitative sequences with limited data: demon co-registration approach
Habib Rebbah1 and Timothé Boutelier1
1Research & Innovation, Olea Medical, La Ciotat, France
1Research & Innovation, Olea Medical, La Ciotat, France
Synopsis
Keywords: Myocardium, Segmentation
Obtaining databases with expert’ manual segmentations to train U-Net is an essential but also a time-consuming operation. We explore here the effect of expanding such a database to other cases with a demon co-registration algorithm. The purpose is to speed up the manual steps but also to allow the development of segmentation algorithms for new cases. We illustrate our approach with MOLLI’ masks expanded to T2 and perfusion data.Introduction
While the U-Net1 architecture became the state of the art approach for automatic fast segmentation in medical image processing, the essential databases with corresponding masks derived by experts are not always available or legally accessible and getting them is not trivial. Indeed, defining the masks is a time-consuming task that must be achieved by specific experts which limits their availability. We propose here to explore the use of demon algorithm2 to obtain masks for multiple sequences.Methods
In the context of the MARVELOUS RHU, we had a database of STEMI patients (HIBISCUS-STEMI, NCT03070496) with MOLLI, T2 prepared (T2p), and perfusion sequences. We also dispose of experts’ masks for MOLLI acquisitions. The aim was to obtain segmentation algorithms of the myocardium based on U-Net for the three sequences.The MOLLI’s algorithm was produced in a classical approach since the corresponding masks were available. This point will then not be explored further in the following. To obtain the masks for T2p and perfusion sequences we used demon algorithm to perform co-registration between MOLLI and these sequences and applied the obtained transformations on the MOLLI’s masks.
The co-registration process is a multi-scale demon algorithm with 3 levels with a shrink factor of 2 between each level and a sigma of the smoothing deformation field of 2.5 voxels and 50 iterations. The demon algorithm requires images with same intensity, which is not the case for the images of the sequences to register. We used a Sobel filter to obtain image descriptors for each sequence and apply the co-registration on them. The MOLLI images and their masks were interpolated to correspond to the resolution of the target sequence images, respectively by linear interpolation and closet point. The sequences processed are time series acquisitions. We choose as images to carry out the co-registrations: the last frame for MOLLI and T2p, and the image with highest standard deviation for perfusion sequence. The series selected for T2p and perfusion sequences correspond to the same patients and to the closest slice location compared to the MOLLI’s ones. The figure 1 illustrates the approach.
The obtained dataset of masks for each sequence were cleaned manually to eliminate the cases of clear failures. The resulting datasets were then used to train the same networks to obtain two segmentation algorithms of the myocardium for T2p and perfusion sequences. The process consists of two U-Net, the first one to detect a bounding box around the myocardium (deduced by the result of the estimated segmentation) while the second one takes as input the cropped image corresponding to the detected bounding box and returns the myocardium segmentation. The division between training and evaluation dataset was 80/20% for both fitting. The figure 2 illustrates the methodology.
In addition to Dice evaluation, a manual comparison between predicted masks and the ones used for the training was executed. Indeed, the co-registration process introduces biases, especially for the perfusion case for which we observe the highest difference of resolution compared with MOLLI sequence. In other term, the masks used for the fitting are considered noised. Figure 3 shows examples of such comparisons.
The U-Net was built using Tensorflow framework and all the developments were done in Python.
Results
The datasets obtained for perfusion and T2p sequences were constituted respectively of 88 and 99 cases. The median Dice value evaluated on the validation part of datasets for perfusion and T2p sequences was respectively 0.75 and 0.81 (figure 4). The predicted masks for perfusion and T2p sequences were considered more accurate than the ones used for the fitting respectively in 87.5% and 58.8% of the cases of evaluation datasets.Discussion
Unlike the experts’ manual segmentations, the obtained ones by co-registration present some thin imperfections (the boundaries do not match perfectly). The U-Net shows a great ability to deal with such uncertainty since it was able to reconstruct the underlying structure of the myocardium and predict segmentations that were more accurate than the proposed ones for the training.The difference of accuracy of the predictions between perfusion and T2p cases highlighted the bias introduced by the co-registration step. Indeed, the latter used an interpolation step in case of perfusion compared with the T2p case which has the same resolution as MOLLI.
The results are encouraging since they demonstrate the feasibility of reducing the time required to produce segmentation databases by experts. However, they also suggest that such an approach should be carried out carefully, considering the specificity of each case (here mainly the difference of resolution between initial and target datasets).
Conclusion
The co-registration allowed to expand an expert segmentation database to other cases while the U-Net was able to manage the uncertainty introduced by such a processing step.Acknowledgements
This work was supported by the RHU MARVELOUS (ANR-16-RHUS-0009). The authors particularly thank Pierre Croisille, Magalie Viallon, Nathan Mewton, Charles De Bourguignon, and Lorena Petrusca for data acquisition and management.References
1. Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 (eds. Navab, N., Hornegger, J., Wells, W. M. & Frangi, A. F.) 234–241 (Springer International Publishing, 2015). doi:10.1007/978-3-319-24574-4_28.
2. Vercauteren, T., Pennec, X., Perchant, A. & Ayache, N. Symmetric Log-Domain Diffeomorphic Registration: A Demons-Based Approach. in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2008 (eds. Metaxas, D., Axel, L., Fichtinger, G. & Székely, G.) 754–761 (Springer, 2008). doi:10.1007/978-3-540-85988-8_90.
Figures
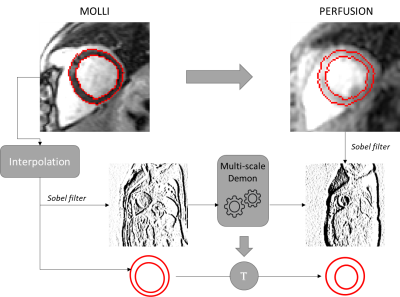
Figure 1 - Illustration
of demon co-registration for expanding segmentation database. Here, from MOLLI to
perfusion cases.
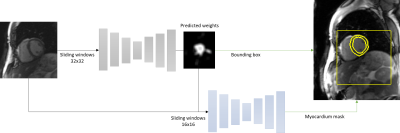
Figure 2 - Illustration of the networks used for myocardium
segmentation. Here with T2p sequences. 2 U-Net: first
for detecting a bounding box around the myocardium; second for fine
classification.
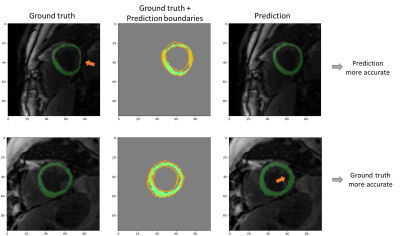
Figure 3 - Illustration
of comparison between predicted segmentations and "ground truth" used
for the fitting process. The first row presents a case for which the prediction
is more accurate: the ground truth does not match the lateral myocardium
boundaries. The second row presents the opposite case: the prediction crosses
the myocardium/left ventricle edge.
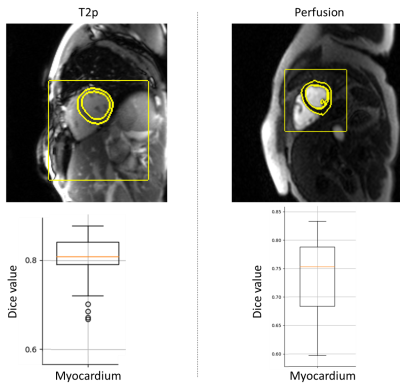
Figure 4 - Results
of fitting process computed with the evaluation datasets for T2p and perfusion
sequences. The two images presents examples with the predicted bounding box and
myocardium mask boundaries.
DOI: https://doi.org/10.58530/2023/4975