4974
Improving cardiac cine MRI using a deep learning-based ESPIRiT reconstruction with self attention
Terrence Jao1, Christopher Sandino2, and Shreyas Vasanawala1
1Department of Radiology, Stanford University, Stanford, CA, United States, 2Department of Electrical Engineering, Stanford University, Stanford, CA, United States
1Department of Radiology, Stanford University, Stanford, CA, United States, 2Department of Electrical Engineering, Stanford University, Stanford, CA, United States
Synopsis
Keywords: Heart, Machine Learning/Artificial Intelligence, Deep Learning, Unrolled, Self Attention, CINE
A deep learning based ESPIRiT (DL-ESPIRiT) was recently proposed to reconstruct dynamic MRI data with higher reconstruction accuracy. However, the method still has difficulty resolving fine anatomic structures. We propose incorporating self-attention to the network using a computationally lightweight squeeze-excitation block (DL-ESPIRiT SE), which uses global information to select more important features while suppressing less important ones. We demonstrate improved reconstruction with DL-ESPIRiT SE, which is most pronounced during faster cardiac motion such as in ventricular ejection.Introduction
Cardiac CINE MRI is widely used for characterizing heart morphology and function, but requires multiple breath holds to minimize motion artifacts from respiration. A deep learning based ESPIRiT (DL-ESPIRiT) reconstruction method was recently proposed to reconstruct dynamic MRI data with up to 12x undersampling and was shown to have higher reconstruction accuracy than state of the art compressed sensing algorithms.1 However, DL-ESPIRiT still has difficulty resolving fast moving, fine anatomic structures such as the papillary muscles. We propose incorporating self-attention into the network to improve the reconstruction fidelity of such structures. Attention works under the premise that certain features of an input or image are more important and relevant than others. Specifically, we incorporate a computationally lightweight architectural unit called a squeeze-excitation block2 into the DL-ESPIRiT network (DL-ESPIRiT SE), which uses global information to self-select more important features and suppress less important ones.Methods
Squeeze-Excitation BlockGiven an intermediate feature map , the squeeze-excitation (SE) block calculates 1D channel weights, $$$W\in\mathbb{R}^{1 \times 1\times 1\times C}$$$ and multiplies them to the feature map as follows:
$$U'=W(U)\times U$$
Where $$$U'$$$ is the final output. The element wise multiplication operator is performed after appropriately broadcasting the channel weights across the spatiotemporal dimensions. $$$W$$$ is calculated by first squeezing spatial information into a channel descriptor through average pooling:
$$z_c=\frac{1}{H \times W \times D} \sum^H_{i=1} \sum^W_{j=1} \sum^D_{k=1} u_c(i,j,k)$$
where $$$z_c$$$ is the squeezed channel descriptor and $$$H \times W \times D$$$ is the spatiotemporal dimensions of the feature maps $$$U=[u_1,u_2,...,u_c]$$$. Subsequently, the excitation network maps the channel descriptor to a learned set of channel weights, $$$W$$$, to capture channel wise dependencies. This excitation step is performed through a bottleneck composed of a dimension reducing fully connected layer with reduction ratio $$$r$$$, a ReLU, and then a fully connected dimension increasing layer followed by a sigmoid activation. The bottleneck is used to limit model complexity and increase model generalization.
$$s=\sigma(F_2\delta(F_1z))$$
where $$$F_1\in\mathbb{R}^{C \times C/r}$$$, is the dimension reducing fully connected layer, $$$F_2\in\mathbb{R}^{C/r \times C}$$$ is the dimension increasing fully connected layer, $$$\delta$$$ is the ReLU function, and $$$\sigma$$$ is the sigmoid activation.
SE blocks were incorporated into every ResNET module as shown in Figure 1 and within the DL-ESPIRiT network as shown in Figure 2. Specifications of the DL-ESPIRiT network itself are explained in more detail within reference 1.
Dataset
Fully sampled 2D cardiac cine datasets were acquired from 22 healthy volunteers on 1.5T and 3.0T GE MRI scanners using a balanced steady state free precession sequence with a 32-channel cardiac coil. Data were acquired in different standard cardiac views including short axis, 2-chamber, 3-chamber, and 4-chamber views. All data were compressed from 32 to 8 virtual coils. The 22 volunteers were divided into three cohorts for training, validation, and testing (10, 2, 10 split). Variable density undersampling masks were applied to simulate 10-15X acceleration across k-t space.
Results
Figures 3 and 4 show representative reconstructions of DL-ESPIRiT, and DL-ESPIRiT SE on retrospectively undersampled cardiac CINE with varying acceleration factors in the short axis and a 3-chamber view respectively. Figure 3 demonstrates that fine papillary muscle structures are reconstructed with higher fidelity with DL-SPIRiT SE even at higher acceleration. The advantage of DL-ESPIRiT SE is most pronounced when motion is greatest during the cardiac cycle. In Figure 4, the anterior wall of the aortic root is blurred during ventricular ejection when reconstructed with DL-ESPIRiT, which is reconstructed with higher fidelity with DL-ESPIRiT SE.Conclusion
In this work, we demonstrate that self-attention using a light-weight squeeze-excitation block can improve the reconstruction quality of cardiac CINE images, which is most pronounced during faster cardiac motion such as in ventricular ejection.Acknowledgements
No acknowledgement found.References
- Sandino, C. M., Lai, P., Vasanawala, S. S., & Cheng, J. Y. (2020). Accelerating cardiac cine MRI using a deep learning‐based ESPIRiT reconstruction. Magnetic Resonance in Medicine, 85(1), 152–167. https://doi.org/10.1002/mrm.28420
- J. Hu, L. Shen and G. Sun,
"Squeeze-and-Excitation Networks," 2018 IEEE/CVF Conference on
Computer Vision and Pattern Recognition, 2018, pp. 7132-7141, doi:
10.1109/CVPR.2018.00745.
Figures
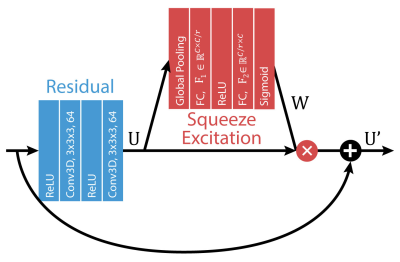
Diagram of the
Squeeze-Excitation block incorporated into a ResNET module. Specifically, the convolutional layers used
in the DL-ESPIRiT network expands the initial images into 64 feature maps.
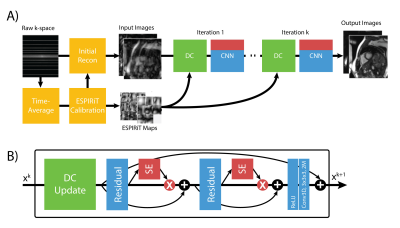
A) The full DLSE-ESPIRiT reconstruction pipeline. ESPIRiT
maps are calculated from a fully sampled calibration region from time averaged
k-space data to compute ESPIRiT maps. These maps are used to calculate initial
zero-filled reconstruction. B) The unrolled network applies data consistency updates
(DC) followed by the squeeze excitation ResNET module.
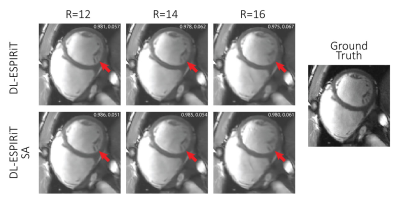
A fully sampled
cardiac CINE dataset in the short axis is undersampled by 12, 14, and 16 during
diastole. Finer structures such as the papillary muscles (red arrow) become
progressively blurrier with higher acceleration. However, DL-ESPIRIT SE is
still able to retain sharpness of this structure even as the acceleration is
increased. Numbers in the top right corner of each image are SSIM, rMSE.
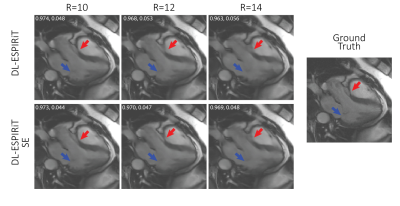
A fully sampled
cardiac CINE dataset in a 3-chamber view is undersampled by 10, 12, and 14
during ventricular ejection in systole. The anterior wall of the aortic root
(red arrow) becomes progressively blurrier with increasing acceleration. The
DL-ESPIRIT SE network retains sharpness of this structure up until an
acceleration of 12. However, neither network is able to recover the mitral
valve leaflets (blue arrow) for this range of acceleration. Numbers in the top left corner of each image are SSIM, rMSE.
DOI: https://doi.org/10.58530/2023/4974