4917
Region of Interest Prediction by Intra-stack Attention Neural Network
Ke Lei1, Ali B. Syed2, Xucheng Zhu3, John M. Pauly1, and Shreyas V. Vasanawala2
1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Radiology, Stanford University, Stanford, CA, United States, 3GE Healthcare, Menlo Park, CA, United States
1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Radiology, Stanford University, Stanford, CA, United States, 3GE Healthcare, Menlo Park, CA, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence, Region of Interest
Manual prescription of the field of view (FOV) by MRI technologists is variable and prolongs the scanning process. Often, the FOV is too large or crops critical anatomy. We propose a deep-learning framework, trained by radiologists’ supervision, for predicting region of interest (ROI) and automating FOV prescription. The proposed ROI prediction model achieves an average IoU of 0.867, significantly better (P<0.05) than two baseline models and not significantly different from a radiologist (P>0.12). The FOV prescribed by the proposed framework achieves an acceptance rate of 92% from an experienced radiologist.INTRODUCTION
At the beginning of a clinical MRI exam, a set of localizer images is collected and MRI technologists will prescribe a FOV for the following scans based on that. This manual step slows down the scanning workflow, prolongs the overall exam time, and influences image quality. Often technologists are not fully informed of radiologist’s requirements to image a region of interest (ROI). Poor FOV prescription may lead to non-diagnostic images, such as relevant anatomy truncated or wrapping artifacts. A conservatively assigned large FOV costs scan time or resolution. Furthermore, studies1-4 have shown that, for longitudinal studies where precise reproducibility of scan prescription is important, automatic methods achieve less variability than manual prescriptions. Therefore, we propose automating the FOV prescription with models trained by radiologists’ ROI labels to get near optimal FOV and streamline scans after the initial localizer.We present a framework where the input is a set of localizer images, the output is a rectangular ROI predicted by a convolutional neural network (CNN) based model, and the final predicted FOV is derived from the ROI according to MRI sampling theory. The high-level workflow is illustrated in Figure 1.
METHODS
The model input is a stack of 2D images and two scalars. We independently train two instances of each model to output a pair of scalars in the range of 0 and 512, representing the top and bottom, or left and right boundaries of the ROI box. Mean-squared error is used as the training loss for all models.Baseline models. We first present two standard CNN models for the ROI regression step.
First is a 2D CNN (Figure 2), referred to as the “2D stacked” model. Slices in a localizer are stacked on the channel dimension. The channel length of the inputs to this model must be constant, so all localizer stacks are zero padded to match the maximum number of slices per stack.
Second is a 3D fully convolutional network (Figure 3), referred to as the “3D” model. This model can take inputs with varying numbers of slices since it is fully convolutional. However, it takes more runtime and memory than the 2D model.
Proposed: shared feature extractor with attention. To get the best of both networks above, we propose using a single-channel-input 2D CNN as the feature extractor shared among all slices, then regress from a linear combination of the extracted features. This architecture allows for a flexible stack size and has less parameters than both networks above.
Furthermore, to obtain an informative combination of slice features, we propose using an attention network to discriminatively weigh the features. The number of slices in our localizer stack ranges from 1 to 40, many of them do not contain relevant anatomy for determining the ROI. A shallow attention network is shared across all slices and trained implicitly within the end-to-end network. Its input is an output of the feature extractor. It outputs a scalar representing the importance of the corresponding slice. The weighted mean of the slice features has a fixed shape regardless of the input stack size. The proposed architecture and training flow are illustrated in Figure 3.
Data augmentation. We create a horizontally flipped copy of all original image stacks. During training, all image stacks are cyclic shifted along the width and height dimensions for a number of pixels randomly chosen from sets {-10,-5,0,5,10} and {-20,-10,0,10,20}, respectively. The boundary labels are adjusted accordingly.
RESULTS
We gathered the axial and coronal view slices in 3-plane localizers from 284 pelvic and 311 abdominal scans. A radiologist labeled one rectangular box on each stack of localizers representing the ground-truth ROI. The coordinates of the box’s four corners are recorded as the labels. The test sets are labeled by two radiologists independently.Quantitative evaluations. ROI predicted by the two baseline models and the proposed model are evaluated by intersection over union (IoU) and position error of the boundaries (in pixel counts out of 512) compared to the ground-truth ROI. The results are shown in Table 1 and 2.
Neither of the two baseline models performs consistently better than the other. The 3D model has four times longer inference time than the 2D stacked and proposed models. The proposed model’s performance is significantly better than both baseline models, and comparable to that of a radiologist.
Reader study. We present the alias-free region using the predicted FOV to a radiologist and ask whether this end result is clinically acceptable. 69 cases get the rating “yes”, 9 cases get the rating “almost”, and 2 cases get the rating “no”.
Figure 4 provides visual examples of the predicted and the labeled ROI.
CONCLUSION
We present a framework for ROI and FOV prediction from MR localizers. The framework utilizes a CNN-based model with intra-stack parameter sharing and attention for ROI prediction, followed by a MR sampling theory derived conversion to FOV. Its performance is significantly better than standard CNN models, and is not significantly inferior to the variance between two radiologists. One limitation of the presented framework is that it does not support oblique ROI and FOV prediction, which will be addressed in future works.Acknowledgements
This work is supported by NIH R01EB009690, NIHR01EB026136, and GE Healthcare.References
- Benner T, Wisco JJ, van der Kouwe AJ, et al. Comparison of manual and automatic section positioning of brain MR images. Radiology. 2006;239(1):246-254. doi:10.1148/radiol.2391050221.
- Kojima S, Hirata M, Shinohara H, Ueno E. Reproducibility of scan prescription in follow-up brain MRI: manual versus automatic determination. Radiol Phys Technol. 2013;6(2):375-384. doi:10.1007/s12194-013-0211-8.
- Itti L, Chang L, Ernst T. Automatic scan prescription for brain MRI. Magn Reson Med. 2001;45(3):486-494.
- Goldenstein J, Schooler J, Crane JC, et al. Prospective image registration for automated scan prescription of follow-up knee images in quantitative studies. Magn Reson Imaging. 2011;29(5):693-700. doi:10.1016/j.mri.2011.02.023.
Figures

Figure 1. Overview of the proposed framework pipeline. First, a CNN based model is trained to predict an ROI for a localizer stack. Then, slice-wise simplified rectangular masks of the object are obtained by setting a threshold on the row or column sums of the pixel values. The final output is the smallest FOV that places aliasing copies of the object masks outside the predicted ROI.
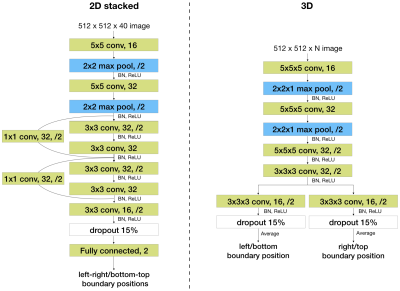
Figure 2. Two baseline network architectures. Network architectures of the 2D stacked baseline model (left) and the 3D baseline model (right). For example, “3x3 conv, 32, /2” represents a convolutional layer with 32 kernels of size 3x3 and using a stride of 2. BN stands for batch normalization. “Fully connected, 2” represents a fully connected layer with two output nodes.
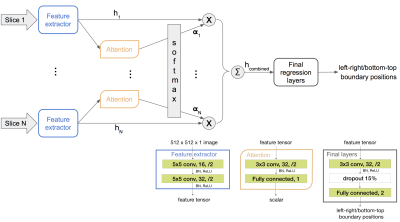
Figure 3. Network flow (top half) and architectures (bottom right corner) of the proposed framework. x represents scaling and Σ represents summation. α are scalars that sum to one. h are image features. The number following “Fully connected” represents the number of output nodes of that layer. There is only one instance of the feature extractor and the attention network, and they are shared across all slices.
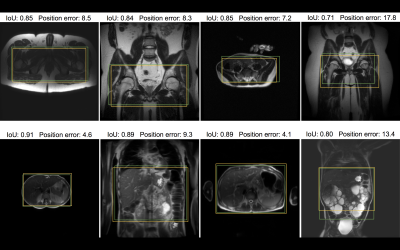
Figure 4. Samples from the test dataset annotated with intra-stack attention model predicted ROI (yellow) and the ROI label given by radiologists (green). On the first row are pelvic localizers, and on the second row are abdominal localizers. Position error and IoU for the model prediction in each example are included to visualize the quantitative results. Note that only one representative slice of a stack is shown, and neither ROI is determined merely from this slice. The bottom right example highlights an extreme case of polycystic kidney disease.
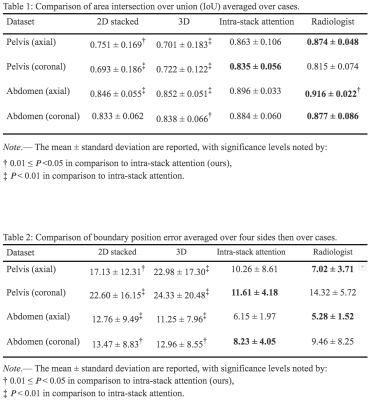
Table 1 and 2. Comparison of area intersection over union (IoU) and boundary position error. Three models are compared with the inter-rater variance between two radiologists across four datasets with 20 cases each.
DOI: https://doi.org/10.58530/2023/4917