4897
In Vivo Single Voxel Relaxation-Diffusion Correlation Spectra Revealed the Microstructure of Human Brain1Department of Integrated Neuroanatomy and Neuroimaging, Kyoto University, Kyoto, Japan, 2Institute of Biophysics, Chinese Academy of Sciences, Beijing, China, 3Human Brain Research Center, Kyoto University, Kyoto, Japan, 4University of Chinese Academy of Sciences, Beijing, China, 5Siemens Shenzhen Magnetic Resonance Ltd, Shenzhen, China, 6Beijing Institute for Brain Disorders, Beijing, China, 7School of Artificial Intelligence, Beijing University of Posts and Telecommunications, Beijing, China
Synopsis
Keywords: Multi-Contrast, Diffusion/other diffusion imaging techniques
Multidimensional MRI resolves the correlations between physical parameters and relaxation properties and has become an increasingly important protocol in biomedical engineering. It has sub-voxel resolution but most research still uses multi-voxel ROIs. In this study, we obtained the single voxel T1-D Relaxation-Diffusion correlation spectra from in vivo human brain tissue boundaries. The significant variance in these spectra might help to identify tissue microstructure at the sub-voxel resolution. Our research disclosed the feasibility of applying multidimensional MRI techniques to detect and segment the human brain.
Introduction
Multidimensional MRI experiments have shown remarkable relevance in resolving the correlation between physical parameters and relaxation properties. The conventional MRI techniques, such as $$$T_1$$$, $$$T_2$$$ and DWI are based on the averaged contrast of different tissues in voxels and thus are not sensitive to microstructures. Compared with these techniques, the multidimensional NMR technique is an effective way to deal with this problem, which allows signal acquisition after multiple evolutions. Thus, it can distinguish overlapping spectral peaks in a correlation spectrum so that the multidimensional NMR technique can obtain the distribution of components within a single voxel and has the potential for sub-voxel imaging. It has become increasingly important in the biomedical field[1], [2]. Multidimensional MRI has suffered from time-consuming and solution accuracy trouble for a long time. Our past research used the genetic algorithm to speed up acquisition and get high-quality solutions which generate multi-voxel ROIs results[3]. Furthermore, we did a normalization and got single-voxel results, which revealed the human brain’s microstructure.Methods
Different from the conventional 1D MRI, the 2D $$$T_1-D$$$ MRI signal takes the general form of [4]: $$ S(TI,b)= \iint F(T_1,D)K_D (b,D) K_(T_1 ) (TI,T_1 )dT_1 dD $$the correlation spectra $$$F(T_1,D)$$$ are calculated by 2D inverse Laplace transform (2D-ILT), which is an ill-conditioned problem owing to the kernel functions’ low-rank features. We used the genetic algorithm (GA) to generate optimized acquisitions based on[3] which could improve accuracy and reduce experimental time.
Two healthy participants were included in the study, and all participants signed informed content approved by the local institutional review board. Experiments were performed on a 3T system (MAGNETOM Prisma-fit, Siemens Healthcare, Erlangen, Germany) equipped with a 20-channel head coil (Nova Medical, Wilmington, MA), and the maximum gradient value was 70 mT/m. Structural images were obtained by T1-weighted magnetization prepared rapid gradient echo (T1w-MPRAGE) with 1.0 mm isotropic resolution.
The IR-DWI images were obtained by the Inversion Recovery pulse prepared Diffusion-Weighted Echo-Planar imaging (IR–DW–EPI) sequence. Note that the inversion times and the b-values are varied in each acquisition while each b-value is in the same direction (Z direction). Certain inversion times and b-values and other parameters are shown in Table 1. Five additional b=0 s/mm2 images were acquired with opposite phase encoding directions for susceptibility distortion and eddy current correction for each inversion time. The total acquisition time was about 50 minutes for each participant.
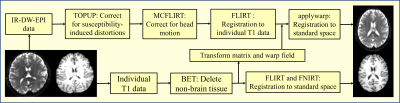
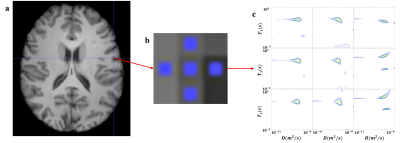
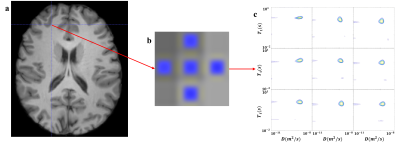
All data preprocessing was carried out using FSL (FMRIB's Software Library) and the detailed pipeline was shown in Figure 1. These steps are referenced to the DWI preprocessing steps. After those procedures, we got normalized IR-DW-EPI and $$$T_1$$$ data and selected single voxel ROIs in the MNI standard space to calculate the spectra. Figures 2a and 3a showed regions of interest (ROI) in standard space, which was manually delineated. We selected nine voxels at the interface of the brain tissues. The signals of these ROIs were inversed with the 2D-ILT software based on [4].
Results and Discussions
In order to detect the microstructure of the human brain, single voxel analysis was performed on the data after normalization from the interface of grey matter and CSF, and white matter and grey matter, as shown in Figures 2a and 3a. As shown in Figures 2b and 3b, conventional $$$T_1$$$ only used voxel averages, which inherently lacked specificity. This led to difficulties in determining the boundaries of different components.With multidimensional MRI, the correlation spectra of components within a single voxel were obtained. As shown in the lower two voxels in the third row of Figure 2c, which were identified as CSF signals on the $$$T_1$$$ image, showed multiple peaks in the 2D spectra. This indicated the existence of components within this voxel other than the CSF. While the spectra for the single voxel at the grey-white matter junction, shown in Figure 3c, did not detect the multiple peaks. The reason was that the grey and white matter tissue was relatively similar in terms of $$$T_1$$$ and diffusivity.
The multidimensional MRI signals’ SNR of the in vivo human is usually not high enough to get accurate spectra after acceleration. That’s the reason why the studies are often qualitative. In our research, we novelly did single voxel research and found multiple peaks in the spectra. Despite its exploratory nature, we cannot classify these peaks as specific microstructures, but this study offered some insight into this technique’s sensitivity to those components.
This technique still needs further consideration, such as more diffusion directions to get the ADC. Further work is required to verify the viability of specific tissue analysis. Generally, our single voxel’s relaxation-diffusion correlation spectra have shown specificity in tissue interface. This technique offers the potential to measure the intra-voxel correlations between the diffusivity and relaxation properties, which may be further applied for precise segmentation and diagnosis of microstructure injuries.
Conclusion
In this study, single voxel’s in vivo $$$T_1$$$ relaxation-diffusion correlation spectra of the human brain were obtained. We achieved pilot research of the sub-voxel imaging by the multi-dimensional MRI. Significant specificity was detected in these spectra, which made a difference in identifying microstructure at the sub-voxel resolution. Our results showed the multidimensional MRI’s potential in precise segmentation and diagnosis of microstructure injuries.Acknowledgements
This work was supported in part by the National Natural Science Foundation of China grant (61901465 and 81871350), Ministry of Science and Technology of China grants (2022ZD0211901, 2020AAA0105601, 2019YFA0707103).
References
[1] D. Benjamini and P. J. Basser, “Multidimensional correlation MRI,” NMR Biomed., vol. 33, no. 12, p. e4226, 2020, doi: 10.1002/nbm.4226.
[2] D. Topgaard, “Diffusion tensor distribution imaging,” NMR Biomed., vol. 32, no. 5, p. e4066, May 2019, doi: 10.1002/nbm.4066.
[3] Lixian Wang et al., “In Vivo Relaxation-Diffusion Correlation revealed MRI Distribution Spectra of the Human Brain Using Genetic Algorithm Optimized Acquisitions,” in In Proceedings of the 31st Annual Meeting of ISMRM, London, England, UK, May 2022, p. Abstract 6263.
[4] Y.-Q. Song, L. Venkataramanan, M. Hurlimann, M. Flaum, P. Frulla, and C. Straley, “T1–T2 Correlation Spectra Obtained Using a Fast Two-Dimensional Laplace Inversion,” J. Magn. Reson. San Diego Calif 1997, vol. 154, pp. 261–8, Mar. 2002, doi: 10.1006/jmre.2001.2474.