4870
Arbitrary Level Contrast Dose Simulation in Brain MRI Using Iterative Reconstruction with Transformer Models.1Subtle Medical Inc., Menlo Park, CA, United States
Synopsis
Keywords: Simulations, Data Acquisition, MRI Dose Simulation
In Gadolinium-based contrast agents (GBCAs) assisted MRI scans, dose simulation is a significant task that enables understanding of the perfusion process. In this work, we propose a new transformer-based iterative model to generate MRI images with arbitrary contrast dosages. By using datasets where only 0%, 10%, and 100% dose images are available, the proposed model can synthesize quantitatively and qualitatively accurate MRI images of varying dosages. The simulated images can be used for other downstream tasks like developing reduced-dose acquisitions and finding the minimum dose needed for a given pathology.Introduction
Gadolinium-based contrast agents (GBCAs) have been widely used in the MRI exams due to their high diagnostic capability. Simulating the images with different contrast dose levels is critical for a better modeling of the contrast perfusion process. Moreover, the MRI images of varying doses can be used for other important downstream applications. For example, the simulated 10% dose image can be utilized to apply contrast-enhancement techniques1,2 when it is impractical to acquire the stipulated low-dose images under varying clinical settings.Currently, arbitrary-dose simulation is being done by physics-based models like the contrast perfusion model3 and contrast quantitative assessment model4. However, there is no deep learning (DL) model along this direction. This is due to the lack of diverse ground truth data of varying low-doses. To the best of our knowledge, this work introduces the first DL model that can synthesize MRI images of arbitrary dosage by training on a highly imbalanced dataset with only 0%, 10%, and 100% dose images. Based on the proposed learning design, we further introduce a new Global transformer (Gformer), in which the self-attention can concentrate more on the global contextual information than conventional approaches.
Methods
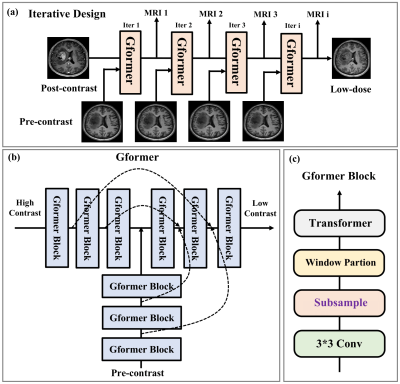
1. Iterative dose simulation learning design:Iterative methods are a traditional strategy to generate stepwise solutions with intermediate outcomes. The proposed iterative model for the dose simulation task is inspired by the state-of-the-art models like LEARN5 and MAPNN6 in CT. As shown in Fig. 1(a). It essentially learns a translation from the post-contrast to the low-dose images with certain iterations. Within each iteration, the higher dose and pre-contrast images are fed to the identical base model to predict the lower dose image. In this manner, all the intermediate outputs correspond to the MRI images of different contrast dosage with a uniform interval.
2. Model convergence:
Iterative models are susceptible to the gradient explosion/vanishing problem, especially when the iteration number grows. To tackle this, we adopt an approach wherein a ‘simulated truth’ is generated by linear interpolation between post-contrast and pre-contrast, serving as fake labels that match the intermediate outputs during training. Specifically, given k iterations, the model generates $$$k$$$ outputs $$$P_i$$$, $$$i = 1, 2 , ..., k$$$. Then, the ‘simulated truth’ $$$S_i$$$, $$$i = 1, 2 , ..., k−1$$$ for iteration $$$i$$$ is calculated as $$S_i = P_r + (P_o-P_r)\times \frac{k-i}{k},$$ where $$$P_r$$$ and $$$P_o$$$ denote the pre-contrast and post-contrast images, respectively. In practice, the simulated truths are assigned only small loss weights for convergence, so they do not overwhelm the loss on the real low-dose image. Specifically, L1 and SSIM losses are calculated on all outputs for model training. In addition, the VGG19-based perceptual loss and the adversarial loss are applied to refine the anatomical textures.
3. Global transformer model (Gformer):
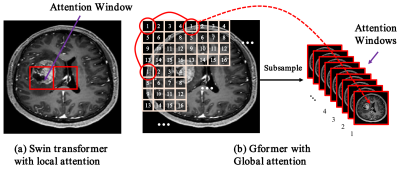
Recently, self-attention-based transformer models have risen to prominence in a wide range of computer vision applications7-14. Inspired by these models, we propose a hybrid global transformer (Gformer) as backbone for dose simulation task. As illustrated in Fig. 1(b), It includes a bi-encoder decoder architecture that takes higher dose and pre-contrast images as input and lower dose images as output. Both encoders and decoder include the proposed Gformer block as a backbone module. As shown in Fig. 1(c), the Gformer block includes a convolution block, a subsample process, a window-partition process, and a typical transformer module. Specifically, the convolution can extract local information while the self-attention will emphasize more the global contextual information. The key element is the subsampling process where we generate 16 sub-images of 64×64 as the attention windows as illustrated in Fig. 2. The Gformer then performs attention on subsampled images that encompass various global contextual information with minimal self-attention overhead on small feature maps.
Results
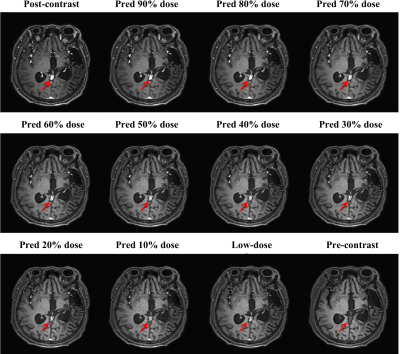
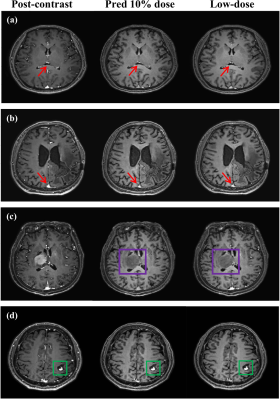
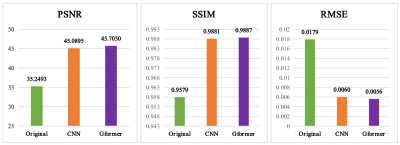
With IRB approval and informed consent, we retrospectively used 125 clinical cases (113 training; 13 testing). For each patient, 3D T1w MPRAGE scans with 240 slices were acquired for the pre-contrast, low-dose, and post-contrast images. These paired images are then normalized and registered during preprocessing. The baseline CNN model is implemented by replacing the Gformer module with convolutional module. Fig. 3 and 4 show the intermediate results and 10% dose predictions, respectively. Fig. 5 shows the results of quantitative assessments between the 10% dose and the low-dose images.Discussion
Fig. 3 shows that the proposed model can generate accurate MRI images that correspond to varying dose levels. Results in Fig. 4 (a) and (b) indicate the high quality of the 10% dose images. Fig. 4(c) shows that the simulated low-dose image’s pathological structure is similar to that of the ground truth. The model’s robustness to ignore contrast-irrelevant enhancement is further illustrated in Fig. 4(d). Finally, quantitative results in Fig. 5 also show that the Gformer achieves superior scores in terms of peak signal-to-noise ratio (PSNR), structural similarity (SSIM), and root mean square error (RMSE).Conclusion
In this work, a Gformer-based iterative model is proposed to simulate the MRI images with arbitrary dosage. The synthesized images can be used for downstream tasks like dose reduction. Furthermore, with arbitrary level doses available, power analysis can be performed to determine the amount of low-dose necessary for each type of pathology.Acknowledgements
We would like to acknowledge the grant support of R44 EB027560.References
Gong E, Pauly J M, Wintermark M, et al. Deep learning enables reduced gadolinium dose for contrast‐enhanced brain MRI[J]. Journal of magnetic resonance imaging, 2018, 48(2): 330-340.
Pasumarthi S, Tamir J I, Christensen S, et al. A generic deep learning model for reduced gadolinium dose in contrast‐enhanced brain MRI[J]. Magnetic Resonance in Medicine, 2021, 86(3): 1687-1700.
Sourbron S P, Buckley D L. Tracer kinetic modeling in MRI: estimating perfusion and capillary permeability[J]. Physics in Medicine & Biology, 2011, 57(2): R1.
Mørkenborg J, Pedersen M, Jensen F T, et al. Quantitative assessment of Gd-DTPA contrast agent from signal enhancement: an in-vitro study[J]. Magnetic resonance imaging, 2003, 21(6): 637-643.
Chen H, Zhang Y, Chen Y, et al. LEARN: Learned experts’ assessment-based reconstruction network for sparse-data CT[J]. IEEE transactions on medical imaging, 2018, 37(6): 1333-1347.
Shan H, Padole A, Homayounieh F, et al. Competitive performance of a modularized deep neural network compared to commercial algorithms for low-dose CT image reconstruction[J]. Nature Machine Intelligence, 2019, 1(6): 269-276.
Shamshad F, Khan S, Zamir S W, et al. Transformers in medical imaging: A survey[J]. arXiv preprint arXiv:2201.09873, 2022.
Liu J, Pasumarthi S, Duffy B, et al. One Model to Synthesize Them All: Multi-contrast Multi-scale Transformer for Missing Data Imputation[J]. arXiv preprint arXiv:2204.13738, 2022.
Wang D, Wu Z, Yu H. Ted-net: Convolution-free t2t vision transformer-based encoder-decoder dilation network for low-dose ct denoising[C]//International Workshop on Machine Learning in Medical Imaging. Springer, Cham, 2021: 416-425.
Liu Z, Lin Y, Cao Y, et al. Swin transformer: Hierarchical vision transformer using shifted windows[C]//Proceedings of the IEEE/CVF International Conference on Computer Vision. 2021: 10012-10022.
Chu X, Tian Z, Wang Y, et al. Twins: Revisiting the design of spatial attention in vision transformers[J]. Advances in Neural Information Processing Systems, 2021, 34: 9355-9366.
Wu S, Wu T, Tan H, et al. Pale transformer: A general vision transformer backbone with pale-shaped attention[C]//Proceedings of the AAAI Conference on Artificial Intelligence. 2022, 36(3): 2731-2739.
Chen C F, Panda R, Fan Q. Regionvit: Regional-to-local attention for vision transformers[J]. arXiv preprint arXiv:2106.02689, 2021.
Ren P, Li C, Wang G, et al. Beyond Fixation: Dynamic Window Visual Transformer[C]//Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2022: 11987-11997.
Figures