4848
Comparison of 17 machine-learning models derived from LGE-MRI for predicting reverse LV remodeling in patients with STEMI
Jianing Cui1, Tao Li1, Xiuzheng Yue2, Sicong Huang2, Yun Kang2, and Fei Yan1
1Radiology, the First Medical center, PLA General Hospital, Beijing, China, 2Philips Healthcare, Beijing, China
1Radiology, the First Medical center, PLA General Hospital, Beijing, China, 2Philips Healthcare, Beijing, China
Synopsis
Keywords: Heart, Heart, reverse left ventricular remodeling
Radiomics is an emerging quantitative imaging method that could extract mineable high-dimensional data from medical images. We investigate the suitable models and significant radiomics features of LGE images in participants with STEMI and assess their value in predicting r-LVR. We chose 17 classification models to analyze all the adiomics features of LGE images in participants with STEMI. Our study found that the model of extra tree classifier was manifested relatively high AUC value in predicting r-LVR. The wavelet-HHH_gldm_SmallDependenceLowGrayLevelEmphasis was relatively strong predictor of r-LVR.Introduction
Over the past few decades, despite the early survival of patients with ST-segment elevation myocardial infarction (STEMI) having improved significantly, many patients are still exposed to a chronic consequence after MI, known as left ventricular (LV) remodeling[1]. Reverse LV remodeling (r-LVR) is the return of the LV geometry to a near-normal elliptical shape[2]. It is associated with the recovery of LV function and improved long-term prognosis[3]. LGE magnetic resonance imaging (MRI) could be used to identify infarcted myocardium qualitatively and quantitatively, contributing valuable information for r-LVR prognostication[4]. Radiomics is an emerging quantitative imaging method proposed in 2012 [5] that could extract mineable high-dimensional data from medical images at the voxel or pixel level. There are 17 machine-learning classifiers used in several clinical studies and they could serve as the bridge between imaging biomarkers and personalized medicine to reflect underlying structural and pathophysiologic information[6]. However, the potential of which machine learning based radiomics analysis with cardiac magnetic resonance (CMR) could be used in prognostication r-LVR remains unknown. The purpose of the current study was to identify the suitable models and significant radiomics features of LGE images in participants with STEMI and assess their value in predicting r-LVR.Materials and Methods
This retrospective study involved 89 STEMI patients who underwent CMR scanning on 1.5-T scanners(Multiva, Philips Healthcare, Netherlands)during the initial week and 5 months after PCI treatment, 38(31.46%) patients with r-LVR. R-LVR was defined as a decreased left ventricular end-systolic volume (LVESV) by 10% or more at the second CMR compared to the first CMR examination [7]. Patients were randomly divided into training (n=69) and validation (n=20) datasets at a ratio of 8:2. Free-hand regions of interest (ROIs) of the global and infarcted myocardium were produced from the basal to the apical slices of the short axial LGE images using the PSIR-LGE sequence. Radiomics features of the two sections, global LV myocardium and infarcted myocardium, were extracted on PSIR-LGE images, respectively. Four types of images are used to perform the feature extraction, which includes shape, original, Laplacian Gaussian transform, and Wavelet transform. The radiomic feature extraction part of the ISMS (Philips) was developed based on the Pyradiomics 3.0.1toolbox(https://pyradiomics.readthedocs.io/en/latest/). A total of 1200 radiomic features were then extracted for each section. To reduce the dimensionality of the dataset, we perform feature selection through five feature selection methods: variance (ANOVA), mutual information (MI), Relief, recursive feature elimination (RFE), and least absolute shrinkage and selection operator (LASSO)). The feature analysis section has a total of 17 classification models, including Logistic Regression, K-Nearest Neighbour Classifier, Parsimonious Bayes, Decision Trees, Gradient Boosting Classifier, Ada Boost Classifier, Light Gradient Boosting Machine, Linear Discriminant Analysis, Extra Trees Classifier, Xgboost. The pipeline of ISMI could calculate the training set and evaluates the performance of all classifier by using 10-fold cross-validation, then choose the top 5 performance model to create the training model. The statistical results include Accuracy, AUC, Recall, and more.Results
For model training in the training dataset, the model with high AUC value was the extra tree classifier (LASSO, infarct myocardium, AUC = 0.68). The highest contributing feature in the model with the highest AUC value was wavelet-HHH_gldm_SmallDependenceLowGrayLevelEmphasis. For model validation in the validation dataset, the accuracy (ACC) value of the extra tree classifier (LASSO, infarct myocardium) was 0.65.Discussion
In our study, the models with high AUC values were random forest classifier (Relief, global myocardium) and extra tree classifier (LASSO, infarct myocardium) in the training dataset. However, the models with highest ACC values were ridge classifier (LASSO, global myocardium), ADA boost classifier (MI, infarcted myocardium), and random forest classifier (MI, infarcted myocardium) (all ACC=0.85). In addition, we considered the values of clinical features, CMR features and laboratory results in predicting r-LVR. Our study found that the model with high ACC value was extra trees classifier in the clinical validation dataset(ACC=0.6). Differences in training and validation of radiomics models, as well as differences between radiomics and clinical models may be related to the amount of data and the sensitivity of the data, which may be improved by incorporating clinical indicators and require further exploration. In summary, we found that the model of extra tree classifier was manifested relatively high AUC value in predicting r-LVR. The wavelet-HHH_gldm_SmallDependenceLowGrayLevelEmphasis was relatively strong predictor of r-LVR.Conclusions
In general, our machine learning models derived from the LGE images could provide accurate predictions of r-LVR after STEMI.Acknowledgements
No acknowledgements in this abstract.References
[1] Zahler D, Lee-Rozenfeld K, Ravid D, et al. (2019) Relation of lowering door-to-balloon time and mortality in ST segment elevation myocardial infarction patients undergoing percutaneous coronary intervention. Clin Res Cardiol 108:1053–1058. [2] Koitabashi N, Kass DA. (2012) Reverse remodeling in heart failure—mechanisms and therapeutic opportunities. Nat Rev Cardiol; 9:147–157 [3] Smiseth OA, T orp H, Opdahl A, et al. (2016) Myocardial strain imaging: how useful is it in clinical decision making? Eur Heart J.;37:1196–1207. [4] Bodi V, Monmeneu JV, Ortiz-Perez JT, et al. (2016) Prediction of Reverse Remodeling at Cardiac MR Imaging Soon after First ST-Segment-Elevation Myocardial Infarction: Results of a Large Prospective Registry. Radiology.;278:54-63. [5] Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [6] Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749-62. [7] Aimo A, Gaggin HK, Barison A, Emdin M, Januzzi JL Jr. (2019) Imaging, Biomarker, and Clinical Predictors of Cardiac Remodeling in Heart Failure With Reduced Ejection Fraction. JACC Heart Fail 7:782-794.Figures
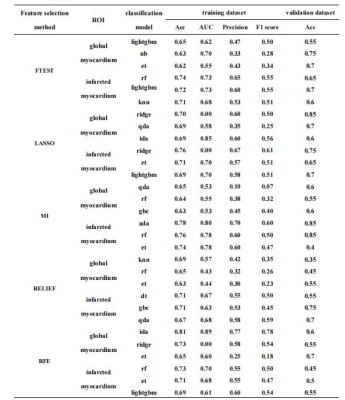
Table 1 The comparison of different classification models in the training and validation datasets.
Lightgbm, Light Gradient Boosting Machine; nb, Naive Bayes; et, Extra Trees Classifier; lr, Logistic Regression; rf, Random Forest Classifier; knn, K Neighbors Classifier; ridge, Ridge Classifier; qda, Quadratic Discriminant Analysis; lda, Linear Discriminant Analysis; gbc, Gradient Boosting Classifier; ada, Ada Boost Classifier; dt, Decision Tree Classifier; lda, Linear Discriminant Analysis.
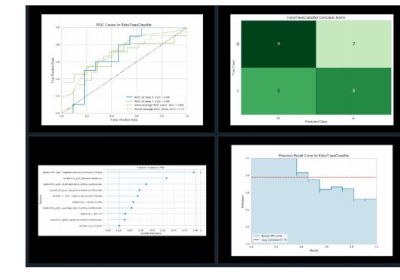
Figure 1 Quantitative assessment of the best models.
DOI: https://doi.org/10.58530/2023/4848