4810
Fast Deep Learning Reconstruction of Interventional MRI Data with Radial, Undersampling k-Space Trajectories1Department of Radiology, University Hospital, LMU Munich, Munich, Germany, 2Department of Statistics, LMU Munich, Munich, Germany, 3Department of Medical Physics, Faculty of Physics, LMU Munich, Munich, Germany
Synopsis
Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence, Radial Acquisition; Undersampled; MR-Guided Interventions
To reduce the data acquisition time and increase frame rates for image guidance during percutaneous needle interventions in the liver, k-space data can be acquired with a radial, undersampling acquisition scheme. The purpose of this work was to optimize a deep learning model for the fast reconstruction of k-space data in this context. The proposed deep learning model reconstructed artificial data with a better image quality compared to conventional reconstruction. Successful reconstruction of interventional phantom data suggests its potential for application during percutaneous needle interventions in the liver.Introduction
Magnetic resonance imaging (MRI) ensures high soft-tissue contrast and does not expose the patient and radiologist to ionizing radiation. This makes MRI superior to other imaging modalities for image guidance during percutaneous needle interventions in the liver1. For interventional procedures, fast data acquisition and reconstruction are inevitable. A reduction of the data acquisition time can be achieved with a radial, undersampling acquisition scheme. However, traditional reconstruction methods for non-Cartesian k-space data, e.g., zero-filling or compressed sensing2 are not ideal for interventional procedures, due to low image quality or high processing times, respectively. In recent years, deep learning-based k-space reconstruction models3,4,5 have been used to reconstruct high quality images while ensuring low inference times. The purpose of this work6 was to optimize a deep learning model for the fast and lightweight reconstruction of radial, undersampled k-space data in the context of MR-guided interventions.Methods
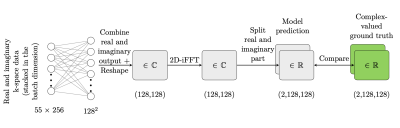
Images from the ImageNet Large Scale Visual Recognition Challenge 2012 (ILSVRC2012) image classification and localization dataset7 were preprocessed to resemble complex-valued MR images. To simulate MR measurements, corresponding radial k-space data with 55 spokes and 256 readouts per spoke were generated using the Python package PyNUFFT8. A fully-connected neural network with no hidden layers (Fig. 1), implemented in PyTorch9, was trained with 100,000 artificial data pairs (radial k-space and complex-valued image) to interpolate radial k-space data to a Cartesian grid. The data were then transformed to the image domain by deploying a 2D inverse fast Fourier transform (iFFT) operation. To increase the robustness of the reconstruction model towards noisy k-space data, 6 (of 55) randomly selected radial spokes were set to zero during training. The deep learning reconstruction was compared to a conventional iterative reconstruction approach using PyNUFFT, which provided a good tradeoff between fast reconstruction time and sufficient image quality. The reconstruction performances of the deep learning and conventional reconstruction were evaluated in terms of image quality quantified as the mean squared error (MSE) and inference time for 50,000 artificial test samples.In the next step, both reconstruction algorithms were tested on radial, undersampled k-space data of a 3D abdominal phantom (triple-modality 3D abdominal phantom, model 057A, Computerized Imaging Reference Systems Inc., Norfolk, USA) acquired with a real-time interactive gradient echo (GRE) sequence on a 1.5-T whole-body MRI system (MAGNETOM Aera, Siemens Healthineers, Erlangen, Germany). The coil measurements were reconstructed individually and then combined with a root-sum-of-squares algorithm.
Results and Discussion
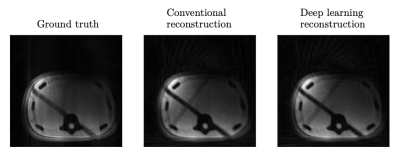
The proposed deep learning model reconstructed artificial k-space data with a better image quality (median [interquartile range] of MSE: 0.0012 [0.0008; 0.0020]) than the conventional iterative reconstruction (0.0018 [0.0012; 0.0026]) (Fig. 2). For the reconstruction of the artificial data with a CPU, the wall-clock inference times for one image were comparable (41[41; 41] ms vs. 44 [43; 44] ms). When performing the calculations on a GPU, the median inference time of the deep learning reconstruction model was reduced to 2 [2; 2] ms.The conventional method and deep learning model achieved comparable reconstruction quality for k-space data from 1.5-T MRI measurements of the interventional abdominal phantom (Fig. 3). This shows that the deep learning reconstruction model, which was trained with artificial data, generalized well for the acquired k-space data.
Conclusion
The proposed deep learning reconstruction model achieved better image quality compared to the conventional iterative reconstruction for the artificial dataset and enabled fast reconstruction times. The successful reconstruction of interventional phantom data showed its potential for application during percutaneous needle interventions in the liver.Acknowledgements
No acknowledgement found.References
1. Fischbach, F., Bunke, J., Thormann, M., Gaffke, G., Jungnickel, K., Smink, J., & Ricke, J. (2011). MR-guided freehand biopsy of liver lesions with fast continuous imaging using a 1.0-T open MRI scanner: experience in 50 patients. Cardiovascular and interventional radiology, 34(1), 188-192.
2. Lustig, M., Donoho, D. L., Santos, J. M., & Pauly, J. M. (2008). Compressed sensing MRI. IEEE signal processing magazine, 25(2), 72-82.
3. Zhu, B., Liu, J. Z., Cauley, S. F., Rosen, B. R., & Rosen, M. S. (2018). Image reconstruction by domain-transform manifold learning. Nature, 555(7697), 487-492.
4. Eo, T., Jun, Y., Kim, T., Jang, J., Lee, H. J., & Hwang, D. (2018). KIKI‐net: cross‐domain convolutional neural networks for reconstructing undersampled magnetic resonance images. Magnetic resonance in medicine, 80(5), 2188-2201.
5. Lin, D. J., Johnson, P. M., Knoll, F., & Lui, Y. W. (2021). Artificial intelligence for MR image reconstruction: an overview for clinicians. Journal of Magnetic Resonance Imaging, 53(4), 1015-1028.
6. Topalis, J. (2022). Fast deep learning reconstruction of interventional MRI data [Unpublished master’s thesis]. Ludwig-Maximilians-Universität München.
7. Russakovsky, O., Deng, J., Su, H., Krause, J., Satheesh, S., Ma, S., ... & Fei-Fei, L. (2015). ImageNet large scale visual recognition challenge. International journal of computer vision, 115(3), 211-252.
8. Lin, J. M. (2018). Python non-uniform fast Fourier transform (PyNUFFT): An accelerated non-Cartesian MRI package on a heterogeneous platform (CPU/GPU). Journal of Imaging, 4(3), 51.
9. Paszke, A., Gross, S., Massa, F., Lerer, A., Bradbury, J., Chanan, G., ... & Chintala, S. (2019). PyTorch: An imperative style, high-performance deep learning library. Advances in neural information processing systems, 32.
Figures