4792
Radial-RIM: accelerated radial 4D MRI using the recurrent inference machine1Netherlands Cancer Institute, Amsterdam, Netherlands, 2Amsterdam UMC location University of Amsterdam, Amsterdam, Netherlands, 3Amsterdam Neuroscience, Amsterdam, Netherlands, 4Oslo University Hospital, Oslo, Norway, 5University of Amsterdam, Amsterdam, Netherlands, 6Radboud University Medical Center, Nijmegen, Netherlands
Synopsis
Keywords: Image Reconstruction, Image Reconstruction, radial, radiotherapy, liver, lung, respiratory motion
To accelerate the acquisition and reconstruction of the respiratory correlated (4D) radial MRI for image guided radiotherapy guidance, we propose a 3D radial-RIM network by adapting the forward model for Cartesian k-space in the regular RIM to radial k-space. Experiments show that the radial-RIM reconstructs images with better quality than the XD-GRASP compressed sensing for 3-4 times accelerated 4D radial free-breathing lung/liver acquisitions, with reconstruction time per slice reduced from 2 minutes to 1.9 seconds. Signal correlation exploited by the radial-RIM over the extra motion-state dimension contributes substantially to the image quality.
Purpose
To accelerate respiratory correlated (4D) radial MRI using the radial recurrent inference machine (radial-RIM).Methods
We propose a 3D radial-RIM network to reconstruct 4D radially sampled k-space by adapting the forward model of the regular RIM1 for the radial acquisition. The network architecture is presented in Figure 1. Validation of the network was performed with reconstruction of free-breathing 3D golden angle stack-of-stars lung and liver scans of healthy volunteers, and with scans of liver metastasis patients, all scanned with the 1.5T Elekta Unity MR-Linac with an 8-channel coil. The sequence parameters are listed in Table 1. Patients were scanned according to the clinical protocol at the Netherlands Cancer Institute, acquiring 1075 k-space spokes with a scan time of 256 seconds. Healthy volunteers were scanned with 1344 spokes with prolonged scan time of 320 seconds for liver and 494 seconds for lung.As preprocessing, we sorted all acquired k-space spokes equally into 10 motion phases2. For each of the sorted 3D radial k-spaces, we applied FFT in the slice direction to obtain the k-space slices. Due to limited GPU memory and the fact that the motion-state dimension is expected to be more redundant than the slice dimension, we focus on the 2D-slice reconstruction along with the extra motion-state dimension. The image size of a slice-over-time is 360x360x10.
The input image for the 3D radial-RIM network was the non-uniform (NU)FFT of the firstly acquired 336 spokes of each scan (i.e., 4x acceleration for the healthy volunteer scans and around 3x acceleration for the liver metastasis patient scans). The compressed sensing (CS) images using XD-GRASP2 with all spokes (CS-full) were regarded as reference in training. In training/validation/testing of the radial-RIM, 7/2/2 subjects of healthy volunteers were used. Each subject includes one liver scan and one lung scan and each scan includes 87 slices. The L1-loss was used in training. For evaluation with the testing data of the healthy volunteer scans, visual inspection was combined with RMSE and SSIM (w.r.t. CS-full reconstruction). The CS reconstruction on the 4x subsampled data (CS-subsampled) was also included as comparison.
To gain insight in the contribution from the extra motion-state dimension, we performed an ablation experiment with a 2D radial-RIM, where the convolutional kernel expanded only in two spatial dimensions and no shared information between motion phases was used.
For evaluation with the scans of the two liver metastasis patients, we performed visual comparison among reconstructions of the 3D radial-RIM and the CS-subsampled with the first 336 k-space spokes, and the CS-full with all acquired 1075 spokes.
Results
The reconstructed images of the test set of the healthy volunteers are shown in Figure 2. Visual inspection shows that the 3D radial-RIM reconstructions largely preserve the visible image features of the reference CS-full reconstructions and meanwhile are less noisy. Note that the CS-full used all spokes and the 3D radial-RIM used a quarter. The reduced number of spokes results in scan time of 124s for lung and 80s for liver.While the CS-subsampled reconstructions are noisier than the CS-full and already show low visibility for small image features, the 2D radial-RIM reconstructions are below the acceptable quality with strong blurriness and artifacts. While the SSIM might be influenced by noise and shows some inconsistency, the RMSE values largely agree to the visual quality.
Figure 3 shows the reconstructed images of the two liver metastasis patients, where the tumor is roughly indicated with the red region of interest. Although the tumor has low visibility with the balanced MRI scan, the 3D radial-RIM shows good agreement on the small image features with the CS reconstructions in the region of tumor and beyond, while being less noisy.
For computation time, the MATLAB implemented CS reconstruction took around 2 minutes per slice (including all motion phases) on the CPU: AMD EPYC 7402 SP3, whereas the PyTorch implemented 3D radial-RIM inference took around 1.9 seconds with the GPU: Nvidia Quadro A6000.
Conclusion
The radial-RIM reconstructs images with better quality than compressed sensing for accelerated 4D radial free-breathing lung/liver acquisitions. The exploited signal correlation over the extra motion-state dimension contributes substantially to the image quality. The shorter acquisition time in combination with radically reduced reconstruction time would allow clinical implementation.Acknowledgements
No acknowledgement found.References
1. Lønning K, Putzky P, Sonke J J, et al. Recurrent inference machines for reconstructing heterogeneous MRI data[J]. Medical Image Analysis, 2019, 53: 64-78.
2. Feng L, Axel L, Chandarana H, et al. XD‐GRASP: golden‐angle radial MRI with reconstruction of extra motion‐state dimensions using compressed sensing[J]. Magnetic Resonance in Medicine, 2016, 75(2): 775-788.
Figures
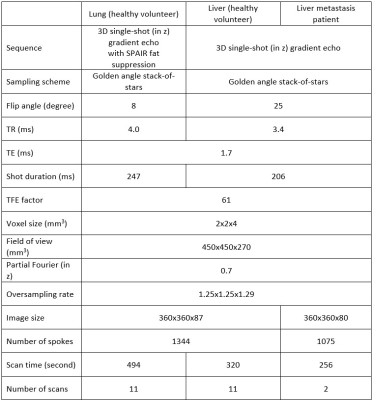
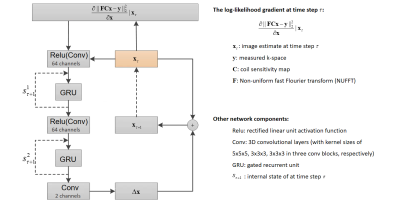
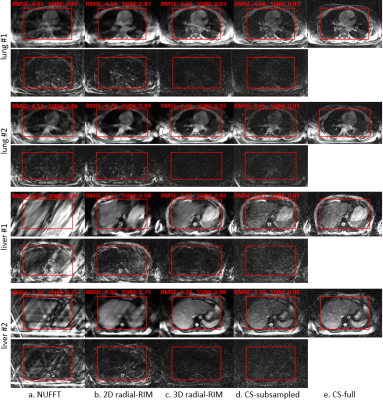
Figure 2. Reconstructed single motion phase of the center slice of the 4D acquisitions of the healthy volunteers with a. NUFFT, b. 2D radial-RIM, c. 3D radial-RIM, d. CS-subsampled and e. CS-full. a-d were reconstructed with the subsampled data (i.e., the first 336 k-space spokes), e with all acquired 1344 spokes. The images are cropped to visualize the region of interest. The RMSE and the SSIM of the 4D volume w.r.t. the CS-full image were computed in the most relevant region inside the red frame.
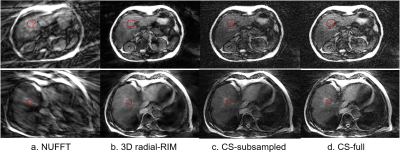
Figure 3. Reconstructed single motion phase of the center slice of the 4D acquisitions of the two liver metastasis patients with a. NUFFT, b. 3D radial-RIM, c. CS-subsampled and d. CS-full. a-c were reconstructed with the subsampled data (i.e., the first 336 k-space spokes), d with all acquired 1075 spokes. Each row represents one patient. The images are cropped to visualize the region of interest. The red region of interest roughly indicates the region of tumor.