4788
MONAI Recon: An Open Source Tool for Deep Learning Based Accelerated MRI Reconstruction1Rice University, Houston, TX, United States, 2NVIDIA, Santa Clara, CA, United States, 3Technical University of Munich, Munich, Germany
Synopsis
Keywords: Software Tools, Machine Learning/Artificial Intelligence, MRI Reconstruction
Deep learning models outperform traditional methods in terms of quality and speed for numerous medical imaging applications. A critical application is the acceleration of magnetic resonance imaging (MRI) reconstruction, where a deep learning model reconstructs a high-quality MR image from a set of undersampled measurements. For this application, we present the MONAI Recon Module to facilitate fast prototyping of deep-learning-based models for MRI reconstruction. Our free and open-source software is pre-equipped with a baseline and a state-of-the-art deep-learning-based reconstruction model and contains the necessary tools to develop new models. The developed open-source software covers the entire MRI reconstruction pipeline.Introduction
Deep learning based reconstruction models are state-of-the-art in accelerated MRI reconstruction. To provide the community with the ability to conduct reproducible research and develop new models in this area, it is highly beneficial to start establishing an open-source foundation. To date, however, there are only a few open-source projects to achieve this goal for accelerated MRI reconstruction [9, 11, 6]. Most of the existing open-source projects are limited to a certain group of neural networks and lack a seamless connection to standard deep learning pipelines (for example, common data processing functionalities or the tools to develop new deep learning models).In this work, we develop the MRI reconstruction module for MONAI (Medical Open Network for AI). MONAI is an open-source project created to establish a community of AI researchers to develop and exchange best practices for AI in healthcare imaging across academia and enterprise researchers.
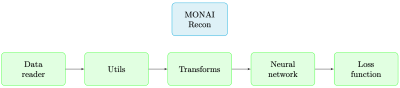
Our MRI reconstruction software, aside from providing standard training and inference features similar to other software, enjoys the following distinctive features:
- Our software is directly connected to a rich set of features from MONAI. This is a competitive advantage since (1) it makes our module inclusive in the sense that every step of the reconstruction task can be done within the MONAI library, and (2) researchers may easily apply ideas from other medical imaging tasks to MRI reconstruction as MONAI supports various medical imaging tasks. For example, MONAI already comes with a bank of network components to develop new neural networks. Or in another example, MONAI is equipped with a large set of spatial, intensity, and frequency-domain transforms created for medical imaging tasks.
- Our reconstruction pipeline (including training and inference) is purely based on PyTorch [5] through MONAI. This provides expert users with more flexible framework and debugging tools as opposed to packages that perform training and inference under the hood via built-in functions (e.g., PyTorch Lightning).
Features
To understand our software’s components, let us start with a formal formulation of the task. Accelerated multi-coil MRI reconstruction deals with reconstructing a ground-truth image $$$\mathbf{x}^*$$$ from a set of $$$k$$$-space measurements $$$\mathbf{y}_1, \ldots , \mathbf{y}_{n_c}$$$ of $$$n_c$$$ receiver coils undersampled with a given mask $$$\mathbf{M}$$$. Given n such samples with paired ground-truth images, we developed a supervised MRI reconstruction pipeline with the following components (also depicted in Figure 2):- Data reader: Our data reader supports loading HDF5 dictionary-style datasets (which is a popular format in the field). For example, in the fastMRI [11] dataset, each sample has the keys {kspace, reconstruction_rss, acquisition, max, norm, patient_id}. In this case, our data reader returns those keys with their corresponding values. Also, note that our constraint is merely on the format of the dataset, and hence any general 2D/3D MRI scan that can be converted to that format can be processed with our software.
- Utilities: Since MRI datasets are typically complex-valued, we implemented several utility functions to handle this particular data type throughout the reconstruction pipeline. Several examples of such functions include complex absolute value, complex multiplication, and complex transformation of an array to a tensor.
- Transforms: In a standard deep learning framework, transform typically refers to an operation performed on the input data of the network (before being fed to the network). For MRI reconstruction, because most of the datasets are fully-sampled, we implemented undersampling transforms to retrospectively undersample the input k-space for the research purpose in simulating compressed sensing MRI. In this regard, we implemented the two popular mask types, namely random and equispaced masking transforms [3, 4]. We also implemented a base class for users to define new arbitrarily-patterned mask transforms. Further, since our software is directly connected to MONAI, users can utilize a vast set of spatial and intensity transforms such as intensity normalization, randomized cropping/padding, etc., to improve the performance of the reconstruction algorithm.
- Reconstruction model: We implemented the state-of-the-art E2E-VarNet [8] in our software. MONAI also contains various neural networks developed for different medical imaging tasks (for example the baseline U-Net [7]). Our software is built on top of the foundation provided by MONAI, and hence users can use MONAI’s ample collection of network components to develop new reconstruction models.
- Loss function / metric: For our software, we implemented the frequently-used SSIM loss function and consequently implemented the SSIM metric as well. SSIM is currently the metric of choice in many of the proposed methods and challenges in the field [11, 10, 2, 1]. Another very popular metric in the literature is peak signal-to-noise ratio (PSNR) which is already incorporated in MONAI.
- Documentation and tutorial: MONAI, including the MONAI Recon Module, contains a detailed documentation for every component. Docstring based descriptions for every function (or class) regrading the input/output arguments and hyper-parameter configurations have also been provided. We also developed two demos to show how one can use the reconstruction pipeline explained above. The first demo is dedicated to the baseline U-Net and the second demo showcases MONAI Recon for E2E-VarNet. One inference example from our tutorials is shown in Figure 3. Training and inference specifications (e.g., inference SSIM and runtime) are also shown in Table 1&2 when using U-Net and E2E-VarNet with our software.
Acknowledgements
The work was mainly done during Mohammad Zalbagi Darestani's internship at NVIDIA Research.
M. Zalbagi Darestani and R. Heckel are (partially) supported by NSF award IIS-1816986, and R. Heckel acknowledges support by the Institute of Advanced Studies at the Technical University of Munich, and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) - 456465471, 464123524.
References
[1] Y. Beauferris, J. Teuwen, D. Karkalousos, N. Moriakov, M. Caan, G. Yiasemis, L. Rodrigues, A. Lopes, H. Pedrini, L. Rittner, et al. “Multi-coil MRI reconstruction challenge—assessing brain MRI reconstruction models and their generalizability to varying coil configurations”. In: Frontiers in Neuroscience. Vol. 16. 2022.
[2] Z. Fabian and M. Soltanolkotabi. “HUMUS-Net: Hybrid unrolled multi-scale network architecture for accelerated MRI reconstruction”. In: Advances in Neural Information Processing Systems. 2022.
[3] F. Knoll, T. Murrell, A. Sriram, N. Yakubova, J. Zbontar, M. Rabbat, A. Defazio, M. J. Muckley, D. K. Sodickson, C. L. Zitnick, et al. “Advancing machine learning for MR image reconstruction with an open competition: Overview of the 2019 fastMRI challenge”. In: Magnetic Resonance in Medicine. 2020.
[4] M. J. Muckley, B. Riemenschneider, A. Radmanesh, S. Kim, G. Jeong, J. Ko, Y. Jun, H. Shin, D. Hwang, M. Mostapha, et al. “State-of-the-art machine learning MRI reconstruction in 2020: Results of the second fastMRI challenge”. In: IEEE Transactions on Medical Imaging. 2021.
[5] A. Paszke, S. Gross, F. Massa, A. Lerer, J. Bradbury, G. Chanan, T. Killeen, Z. Lin, N. Gimelshein, L. Antiga, et al. “PyTorch: An imperative style, high-performance deep learning library”. In: Advances in Neural Information Processing Systems (NeurIPS). Vol. 32. 2019.
[6] Z. Ramzi, P. Ciuciu, and J. L. Starck. “Benchmarking deep nets MRI reconstruction models on the fastMRI publicly available dataset”. In: IEEE International Symposium on Biomedical Imaging. 2020, pp. 1441–1445.
[7] O. Ronneberger, P. Fischer, and T. Brox. “U-Net: convolutional networks for biomedical image segmentation”. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. 2015, pp. 234–241.
[8] A. Sriram, J. Zbontar, T. Murrell, A. Defazio, C. L. Zitnick, N. Yakubova, F. Knoll, and P. Johnson. “End-to-end variational networks for accelerated MRI reconstruction”. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. 2020, pp. 64–73.
[9] M. Uecker, J. Tamir, F. Ong, and M. Lustig. “The BART toolbox for computational magnetic resonance imaging”. In: International Society for Magnetic Resonance in Medicine (ISMRM). Vol. 24. 2016.
[10] G. Yiasemis, J. J. Sonke, C. Sánchez, and J. Teuwen. “Recurrent variational network: A deep learning inverse problem solver applied to the task of accelerated MRI reconstruction”. In: IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2022, pp. 732–741.
[11] J. Zbontar, F. Knoll, A. Sriram, M. J. Muckley, M. Bruno, A. Defazio, M. Parente, K. J. Geras, J. Katsnelson, H. Chandarana, et al. “fastMRI: An open dataset and benchmarks for accelerated MRI”. In: Radiology: Artificial Intelligence. 2020.
Figures
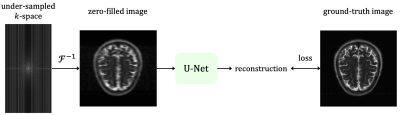
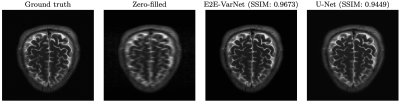

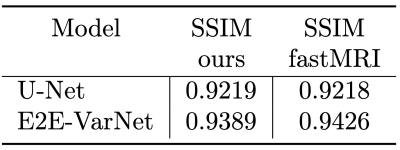
Our software successfully reproduces the results of original implementations of the baseline U-Net and the state-of-the-art E2E-VarNet models. SSIM is for 8× accelerated MRI reconstruction for the brain leaderboard of the fastMRI dataset. For more results, please visit our tutorial page at
https://github.com/Project-MONAI/tutorials/tree/main/reconstruction/MRI_reconstruction.