4785
Lightweight encoder-decoder architecture for Fat-Water separation in MRI using biophysical model-guided deep learning method1Centre for Biomedical Engineering, Indian Institute of Technology Delhi, New Delhi, India, 2Department of Radio Diagnosis, All India Institute of Medical Sciences, New Delhi, India, 3Department of Biomedical Engineering, All India Institute of Medical Sciences, New Delhi, India
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction, Deep Learning, Fat-Water seperation
In this study, we propose a novel lightweight encoder-decoder architecture for deep learning based Fat-Water separation in multi-echo MRI data. The architecture's performance is evaluated in the biophysical model-guided deep learning-based Fat-Water separation task and compared against the widely used U-Net. This biophysical model-guided deep learning-based Fat-Water separation requires no training data and ground truths, but it involves time-consuming loss minimization for thousands of epochs. Despite having significantly fewer training parameters, the proposed architecture performed equally well in generating the Fat-Water maps compared to the U-Net. So, our proposed architecture aids in the faster generation of Fat-Water maps.Introduction
Separating the Magnetic Resonance Imaging (MRI) signal from Fat and Water protons generating the individual Fat and Water maps of any region of interest in the human body is of great clinical significance1. These Fat maps generated can quantify the amount of fat deposited and this quantified metric acts as a biomarker for multiple diseases like Non-alcoholic fatty liver, cardiovascular diseases, etc. The usage of deep learning (DL)-based methods for the Fat-Water separation task has produced noise- and artifact-free maps compared to the traditional algorithms2-6. However, most of the methods proposed in the literature using DL-based Fat-Water separation2-6 utilize a U-Net or other computationally heavy encoder-decoder architectures. In this study, we propose a novel lightweight encoder-decoder architecture for DL-based Fat-Water separation and investigate its performance. The proposed architecture has significantly fewer training parameters than the widely used U-Net. Thus, it is computationally less expensive and faster to reconstruct the multi-echo MRI data, generating the Fat-Water maps.Methods
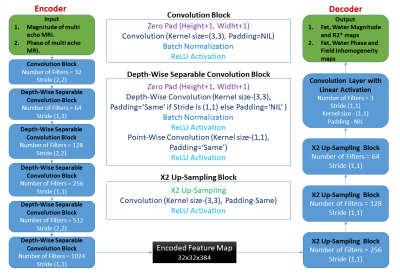
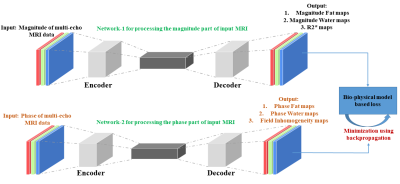
Lightweight encoder-decoder architecture:Figure 1 illustrates the proposed novel lightweight encoder-decoder architecture for Fat-Water separation in multi-echo MRI data using deep learning. The architecture utilizes depth-wise separable convolutions in the encoder instead of traditional convolutions. Depth-wise separable convolutions are computationally efficient, having significantly fewer training parameters than traditional convolutions7. The decoder's end is added with a convolutional layer with three filters and a linear activation, which produces three output maps. As shown in Figure 2, the magnitude and the phase of input multi-echo MRI data are processed separately with two different networks of the same proposed architecture described in Figure 1. The first network (the magnitude network) takes in only the magnitude of the input MRI data and generates the Fat and Water magnitude maps along with R2* maps. The second network (the phase network) takes only the phase of input MRI data and generates the Fat and Water phase maps along with field inhomogeneity maps.
DL-based ad hoc reconstruction of multi-echo data for Fat-Water separation:
Jafari et al.6 proposed a biophysical model-guided DL method that directly reconstructs a single patient’s multi-echo MRI data, generating Fat-Water maps. This ad-hoc reconstruction method requires no prior training of the deep learning model using a large dataset and its corresponding ground truths. The biophysical model used to estimate the Fat-Water maps from the multi-echo MRI data is presented in by Equation (1):
$$\begin{gathered}I_n(x, y)=\left(\rho_w(x, y)+\rho_f(x, y) \cdot \exp \left(j 2 \pi f t_n\right)\right) \\\cdot \exp \left(j 2 \pi \psi(x, y) t_n\right) \cdot \exp \left(-R_2^*(x, y) t_n\right)\end{gathered}\tag{1}$$
$$$I_n(x, y)$$$ is the signal from the $$$n$$$th echo image at any spatial position $$$(x, y)$$$. $$$\rho_w$$$ is the signal from water protons, $$$\rho_f$$$ is the signal from fat protons, $$$f$$$ is the chemical‐shift frequency between fat and water protons, $$$t_n$$$ is the Time for echo of the $$$n$$$th echo, $$$\psi$$$ is the field inhomogeneity and $$$R_2^*$$$ is the inverse of $$$T_2^*$$$.
Equation (1) is used in place of the ground truth Fat quantification, Water, R2* and field inhomogeneity maps to compute the deep learning model’s loss. This biophysical model loss is minimized by back-propagating for 10,000 epochs with a batch size of 10 and a learning rate of 6x10-4, producing the Fat, Water, R2* and field inhomogeneity maps.
Performance Analysis:
To analyze the efficacy of the proposed architecture, we used it to reconstruct a total of 48 slices of physical Fat-Water phantoms (hosted by Hernando et al.8) and 60 slices of human abdomen multi-echo MRI data (hosted in the 2012 ISMRM Fat-Water separation workshop9). The data was split into batches and reconstructed using the ad-hoc method explained in above section. This same testing data was also reconstructed using the U-Net architecture for performance comparison. The Fat-Water maps obtained using both architectures were compared against the ground truth Fat-Water maps by computing the Structural Similarity Index Measure (SSIM)10. The ground truth Fat-Water maps were generated using the traditional hierarchical IDEAL method proposed by Jiang et al.11, the code available in the ISMRM Fat-Water separation toolbox9.
Results
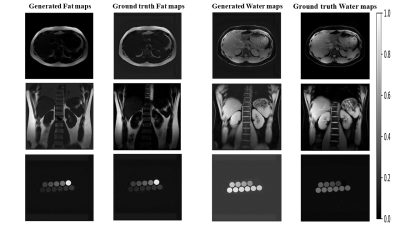
Figure 3 presents the representative sample Fat-water maps generated using the proposed architecture and their corresponding ground truths. Table 1 compares the proposed architecture's and U-Net's performance in the Fat-Water separation task. The proposed architecture achieved an overall average SSIM of 0.837±0.090 and 0.833±0.139 in generating Fat and Water maps, respectively. The U-Net architecture achieved an overall average SSIM of 0.823±0.097 and 0.790±0.182 in generating Fat and Water maps, respectively.Discussion and Conclusion
From Table 1, it is seen that the proposed architecture performed equally well compared to the U-Net architecture. The proposed architecture has only 6,36,683 training parameters, whereas the U-Net has 1,182,579. The ad hoc reconstruction method has the advantage of being well-generalized in reconstructing MRI data with different echo times and acquisition parameters from different regions of interest in the body and their views6. The ad hoc reconstruction method has absolutely no bias due to the restrictions in the dataset used for training the deep learning model. However, it involves time-consuming loss minimization for thousands of epochs. So, our the proposed computationally light encoder-decoder architecture would be highly beneficial to this ad hoc reconstruction method for faster Fat-Water separation.Acknowledgements
The physical Fat-Water phantom multi-echo MRI data utilized in this study was hosted publicly by Hernando et al.8 at (http://dx.doi.org/10.5281/zenodo.48266). The human abdomen multi-echo MRI data utilized in this study was hosted in the 2012 ISMRM Fat-Water separation workshop9.References
1. Thomas, E.L., Fitzpatrick, J.A., Malik, S.J., Taylor-Robinson, S.D. and Bell, J.D., 2013. Whole body fat: content and distribution. Progress in nuclear magnetic resonance spectroscopy, 73, pp.56-80.
2. Cho, J. and Park, H., 2019. Robust water–fat separation for multi‐echo gradient‐recalled echo sequence using convolutional neural network. Magnetic Resonance in Medicine, 82(1), pp.476-484.
3. Goldfarb, J.W., Craft, J. and Cao, J.J., 2019. Water–fat separation and parameter mapping in cardiac MRI via deep learning with a convolutional neural network. Journal of Magnetic Resonance Imaging, 50(2), pp.655-665.
4. Andersson, J., Ahlström, H. and Kullberg, J., 2019. Separation of water and fat signal in whole‐body gradient echo scans using convolutional neural networks. Magnetic resonance in medicine, 82(3), pp.1177-1186.
5. Basty, N., Thanaj, M., Cule, M., Sorokin, E.P., Liu, Y., Bell, J.D., Thomas, E.L. and Whitcher, B., 2021. Swap-Free Fat-Water Separation in Dixon MRI using Conditional Generative Adversarial Networks. arXiv preprint arXiv:2107.14175.
6. Jafari, R., Spincemaille, P., Zhang, J., Nguyen, T.D., Luo, X., Cho, J., Margolis, D., Prince, M.R. and Wang, Y., 2021. Deep neural network for water/fat separation: supervised training, unsupervised training, and no training. Magnetic resonance in medicine, 85(4), pp.2263-2277.
7. Chollet, F., 2017. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 1251-1258).
8. Hernando, D., Sharma, S.D., Kramer, H. and Reeder, S.B., 2014. On the confounding effect of temperature on chemical shift‐encoded fat quantification. Magnetic resonance in medicine, 72(2), pp.464-470.
9. Hu, H.H., Börnert, P., Hernando, D., Kellman, P., Ma, J., Reeder, S. and Sirlin, C., 2012. ISMRM workshop on fat–water separation: insights, applications and progress in MRI. Magnetic resonance in medicine, 68(2), pp.378-388.
10. Zhou Wang, A. C. Bovik, H. R. Sheikh and E. P. Simoncelli, "Image quality assessment: from error visibility to structural similarity," in IEEE Transactions on Image Processing, vol. 13, no. 4, pp. 600-612, April 2004, doi: 10.1109/TIP.2003.819861.
11. Jiang, Y. and Tsao, J., 2012. Fast and robust separation of multiple chemical species from arbitrary echo times with complete immunity to phase wrapping. In Proceedings of the 20th Annual Meeting of ISMRM (Vol. 388)
Figures