4781
Deep Attention Unfolding CNN Architecture for Parallel MRI1Electrical and Computer Engineering, COMSATS University Islamabad, Islamabad, Pakistan, 2Electrical Engineering, University of Poonch Rawalakot, Rawalakot AJK, Pakistan, 3Center for Medical Image Science and Visualization, Linköping University, Linköping, Sweden
Synopsis
Keywords: Image Reconstruction, Artifacts, Parallel MRI, Deep Learning, Attention Mechanism
Deep learning has made significant progress in recent years, however the local receptive field in CNN raises questions about signal synthesis and artefact compensation. This paper proposes a deep learning Spatial-Channel Attention U-Net (SCA-U-Net) to solve the folding problems arising in MR images because of undersampling. The main idea is to enhance local features in the images and restrain the irrelevant features at the spatial and channel levels. The output from SCA-U-Net is further refined by adding a small number of originally acquired low-frequency k-space data. Experimental results show a better performance of the SCA-U-Net model than classical U-Net model.Introduction
Magnetic Resonance Imaging (MRI) is a demanded medical imaging technique because it provides high spatial resolution and fine contrast images of human organs. The prolonged scan time of MRI is one of its primary drawbacks. Undersampled measurements in Parallel MRI decrease scan time; however folding artefacts appear as a result.Several deep-learning methods have been proposed in the literature to solve the folding problems in MR images. For instance, it is suggested to estimate the folding artefacts in aliased images using residual convolutional neural networks (CNN)1. Also, CNN followed by the addition of a small number of low-frequency k-space data (originally acquired) has been proposed to solve the folding problems in MR images2. Further, end-to-end fine-tuning has been used for CNN to solve the folding problems and data scarcity issues in MR images3.
U-Net is a CNN encoder and decoder network4 that offers good learning capability with a small training data set. U-Net attains high performance in learning because of the efficient extraction of the local image features4. However, the convolution operators in U-Net have local receptive fields, so the processing of long-range dependencies goes through multiple convolutional layers, which may prevent learning about long-term dependencies5; and can hinder in integrating signals from a broad range of inputs.
One way to extend the receptive field of convolution operators is by employing a hierarchical network architecture6. In fact, integrating CNN with the attention mechanism7, which would detect long-range dependencies throughout the image region, has become popular. For example, the self-attention mechanism in CNN has been proposed for a reliable MR image reconstruction8. Sanghyun et.al9 introduced Convolutional-Block-Attention-Module (CBAM) to detect the long-range dependencies across the image region.
In this research, we propose additional channel and spatial attention mechanisms integrated into CNN (U-Net) for improved MR image reconstruction.
Method
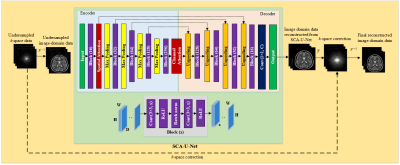
In the proposed method i.e. SCA-U-Net, Spatial and Channel Attention mechanisms9 have been integrated into the U-Net architecture as shown in Fig.1.The acquired undersampled data is initially preprocessed to produce zero-padded data by first adding zeros in the unmeasured areas. Next, the folded image is obtained by taking the inverse Fourier Transform (FT) (absolute value); and it is fed into the trained SCA-U-net to generate the output.
In SCA-U-Net, the encoder part encodes the input image into features at multiple different levels. The role of the decoder part is to gradually restore the details and spatial dimensions of the image according to the image features; as well as the fusion of high-level and low-level features. In addition, spatial attention and channel attention modules are integrated into U-Net as shown in red blocks in Fig.1.
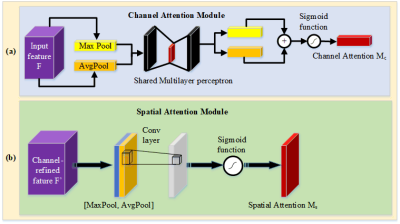
The spatial attention module is added to the low-level feature maps since it mainly extracts the spatial features such as contours and edges. Similarly, the channel attention module is added as the last layer of the encoder, since the high-level feature maps mainly express complex features with a large receptive field and more channels. This mechanism allows the network to perform feature recalibration, through learning to exploit global information to selectively enhance useful features and restrain useless features. The structures of the channel attention module and spatial attention module as used in this paper are shown in Fig. 2.
The final output from SCA-U-Net is converted into k-space data by applying FT; a small number of low-frequency k-space data points are added from the originally measured k-space data to solve the localization uncertainty due to image folding. Finally, the solution image is obtained by applying inverse FT.
Results and Discussion
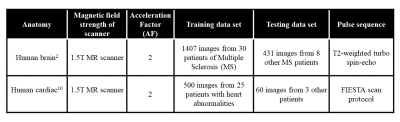
The details of training and test datasets are given in Table 1.Training of the proposed SCA-U-Net involves the input and output images that are pairs of the FT of the subsampled and fully-sampled k-space data. The uniformly undersampled image domain human brain data set at Acceleration Factor (AF) = 2 is used as an input and the corresponding fully-sampled human brain data set is used as an output for training the network in our experiments.
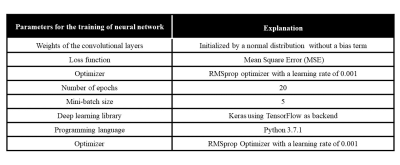
In this paper, the training of the proposed SCA-U-Net is performed on Intel(R) core (TM) i7-4790 CPU, clock frequency 3.6GHz, 16 GB RAM, and GPU NVIDIA GeForce GTX 780 for approximately 3 hours. The libraries and hyper-parameters tuned for the network are shown in Table 2.
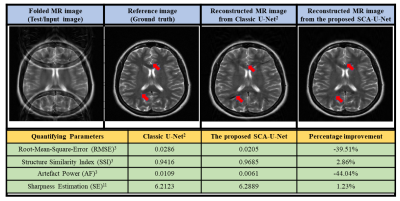
The reconstruction results obtained from the proposed SCA-U-Net are compared with the classic U-Net2. The results are compared visually and in terms of different quantifying parameters i.e. Root-Mean-Square-Error (RMSE), Structured-Similarity-Index (SSI), Artefact-Power (AP)3 and image sharpness estimation11 as shown in Fig.3.
The results show that the proposed SCA-U-Net provides significantly better reconstruction results as compared to the classic U-Net2.
Conclusion
The proposed method i.e. SCA-U-Net improves the reconstruction outcome by taking the advantage of long-range dependencies across the image region. The improved image fidelity for uniformly undersampled MR images comes by incorporating the attention mechanism into U-Net. In future, the generalization capability of the trained SCA-U-Net can be analyzed for different anatomies and acceleration factors.Acknowledgements
I am extremely obliged to my Ph.D. research supervisor Dr. Hammad Omer (Medical Image Processing Research Group (MIPRG) leader) for his constant guidance, positive denunciation, and cordial assistance at each stage of research. I would like to thank MIPRG, COMSATS University Islamabad, Pakistan, for providing me a superb environment and computational resources for my research work. I am also thankful to ISMRM for providing excellent platform for young researchers doing research in the field of Magnetic Resonance Imaging (MRI).References
1. Z. Li et al., "Triple-D network for efficient undersampled magnetic resonance images reconstruction," Magnetic resonance imaging, vol. 77, pp. 44-56, 2021.
2. C. M. Hyun, H. P. Kim, S. M. Lee, S. Lee, and J. K. Seo, "Deep learning for undersampled MRI reconstruction," Physics in Medicine & Biology, vol. 63, no. 13, p. 135007, 2018.
3. M. Arshad, M. Qureshi, O. Inam, and H. Omer, "Transfer learning in deep neural network based under-sampled MR image reconstruction," Magnetic Resonance Imaging, vol. 76, pp. 96-107, 2021.
4. O. Ronneberger, P. Fischer, and T. Brox, "U-net: Convolutional networks for biomedical image segmentation," in International Conference on Medical image computing and computer-assisted intervention, 2015: Springer, pp. 234-241.
5. Y. Lu, Y. Zhao, X. Chen, and X. Guo, "A Novel U-Net Based Deep Learning Method for 3D Cardiovascular MRI Segmentation," Computational Intelligence and Neuroscience, vol. 2022, 2022.
6. M. Z. Alom, M. Hasan, C. Yakopcic, T. M. Taha, and V. K. Asari, "Recurrent residual convolutional neural network based on u-net (r2u-net) for medical image segmentation," arXiv preprint arXiv:1802.06955, 2018.
7. J.-B. Cordonnier, A. Loukas, and M. Jaggi, "On the relationship between self-attention and convolutional layers," arXiv preprint arXiv:1911.03584, 2019.
8. Y. Wu, Y. Ma, J. Liu, J. Du, and L. Xing, "Self-attention convolutional neural network for improved MR image reconstruction," Information sciences, vol. 490, pp. 317-328, 2019.
9. S. Woo, J. Park, J.-Y. Lee, and I. S. Kweon, "Cbam: Convolutional block attention module," in Proceedings of the European conference on computer vision (ECCV), 2018, pp. 3-19.
10. A. Andreopoulos and J. K. Tsotsos, "Efficient and generalizable statistical models of shape and appearance for analysis of cardiac MRI," Medical image analysis, vol. 12, no. 3, pp. 335-357, 2008.
11. C. Feichtenhofer, H. Fassold, and P. Schallauer, "A perceptual image sharpness metric based on local edge gradient analysis," IEEE Signal Processing Letters, vol. 20, no. 4, pp. 379-382, 2013.
Figures