4774
Deep Learning Prediction of Multi-channel ESPIRiT Maps for Calibrationless MR Image Reconstruction1Laboratory of Biomedical Imaging and Signal Processing, the University of Hong Kong, HongKong, China, 2Department of Electrical and Electronic Engineering, the University of Hong Kong, HongKong, China, 3Department of Electrical and Electronic Engineering, Southern University of Science and Technology, Shenzhen, China
Synopsis
Keywords: Parallel Imaging, Data Acquisition, Brain reconstruction, Cardiac reconstruction
We present a U-Net based deep learning model to estimate the multi-channel ESPIRiT maps directly from uniformly-undersampled multi-channel multi-slice MR data. The model is trained with a hybrid loss function using fully-sampled multi-slice axial brain datasets from the same MR receiving coil system. The proposed model robustly predicted ESPIRiT maps from uniformly-undersampled k-space brain and cardiac MR data, yielding highly comparable performance to reconstruction using to acquired reference ESPIRiT maps. Our proposed method presents a general strategy for calibrationless parallel imaging reconstruction through learning from coil and protocol specific data.Introduction
Most existing methods for parallel image reconstruction in image, k-space or hybrid space require coil sensitivity calibration data either from additional pre-scan or autocalibration signals. Often there exists inconsistency between calibration data and undersampled data, e.g., due to motions, causing artifacts in the reconstructed images1,2. ESPIRiT3 is an effective hybrid-space reconstruction method. It utilizes k-space kernel operations to derive a set of eigenvector maps, i.e., ESPIRiT maps, to effectively represent coil sensitivity information. We present a deep learning model to estimate the multi-channel ESPIRiT maps directly from uniformly-undersampled multi-channel multi-slice MR data. The model is trained using fully-sampled multi-slice axial brain datasets from the same MR receiving coil system.Methods
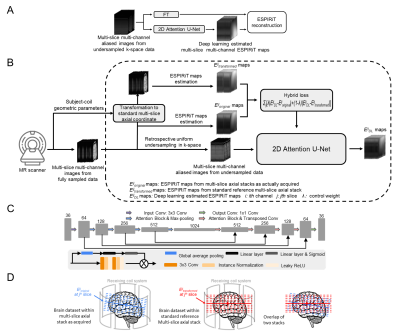
Specifically, a deep learning model is developed for mapping aliased images to the corresponding ESPIRiT maps (Figure 1). The input of the model is the multi-channel aliased MR images from uniformly-undersampled multi-channel data, while the output is the corresponding multi-channel ESPIRiT maps. An attention U-Net model4-6 is adopted (Figure 1C).The coil sensitivity information in any MRI system is largely coil-specific. When scanning a particular subject in the clinical MRI setting, the exact coil sensitivity profiles or ESPIRiT maps within any imaging slice also depend on the orientation/position of the slice with respect to MR receiving coil system (Figure 1D). Such subject-coil geometry information is available during the scan and recorded in the DICOM header. We incorporate such information by imposing a hybrid loss function. Specifically, the model is trained by minimizing a hybrid L1 loss on two sets of multi-slice multi-channel ESPIRiT maps in the following Equation (1). $$ argmin_{\theta} \sum_{ij} [\lambda |E^{ij}_{DL}-E^{ij}_{original}|+(1-\lambda)|E^{ij}_{DL}-E^{ij}_{transformed}|] $$ Here λ is a learnable parameter to control the loss contributions. Specifically, Eijoriginal and Eijtransformed represent two ESPIRiT maps for the ith channel at the jth slice within their original multi-slice locations and their transformed locations within the standard reference multi-slice axial stack (Figure 1D, respectively. This reference stack has a fixed orientation and position relative to the magnet and gradient coil center. Its orientation and position typically have a fixed geometric relation to the coil system. Thus Eijtransformed should be mostly coil specific and dataset independent. Meanwhile, Eijoriginal will be dataset dependent since each multi-slice axial head scan can be prescribed with a slightly different geometry. In practice, Eijtransformed and Eijoriginal differ from each other in position and orientation but will be very similar to a certain extent due to their geometric proximity and the spatial smoothness nature of ESPIRiT maps. Therefore, incorporating Eijtransformed as part of the loss function will indirectly facilitate the learning process through improving stabilization and convergency.
The training and testing data were from the publicly available Calgary-Campinas MR brain database7. They included 65 fully-sampled human brain datasets from 65 individual healthy subjects that were acquired on a 1.5T clinical GE scanner. To further evaluate the robustness of the proposed method, cardiac MRI data from OCMR public database8 were also used. Adam optimizer9 was carried out for training with β1 = 0.9, β2 = 0.999 and initial learning rate = 0.0001. λ was initialized to 0.5 and gradually decreased to zero during training. The training was conducted on a Geforce RTX 3090 GPU using PyTorch 1.8.1 package10 with a batch size of 32 and 100 epochs. The total training time was approximately 27.8 hrs and 7.4 hrs, respectively for the brain 6-channel model and the cardiac 6-channel model. For evaluation, we compared the deep learning results with these using the reference maps derived from 24 consecutive central k-space lines.
Results
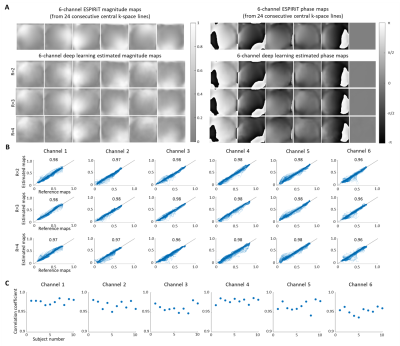
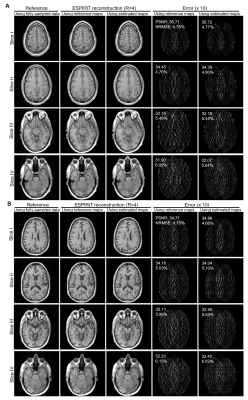
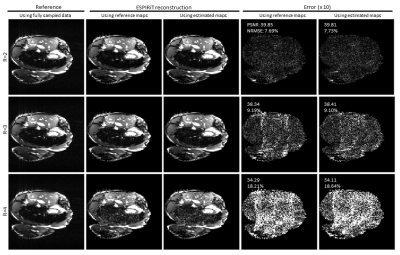
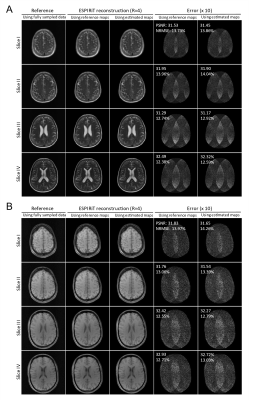
Figure 2 shows the typical results of deep learning estimated ESPIRiT maps. Quantitatively, there existed a high degree of pixel-wise correlation between the deep learning estimated and reference ESPIRiT magnitude maps, indicating the robustness of the proposed deep learning estimation of ESPIRiT maps. Figure 3 presents the results at R = 4 for two subjects with large roll rotation and large overall translation, respectively, again showing similar performance at all slice locations. Figure 4 shows the cardiac results, again demonstrating comparable performance to that using reference maps. These results demonstrated that our method could adapt to the reconstruction of cardiac data, where there are often signal voids and relatively more rapid coil sensitivity variations when compared to brain data.Discussion and Conclusions
The proposed deep learning model is capable of robustly predicting multi-channel ESPIRiT maps from uniformly-undersampled k-space data even at high acceleration. The model training and application are coil specific. However, this may not pose a severe restriction as we envision that, in an era of data-driven computing, truly effective MRI scanners should move towards self-learning, i.e., constant performance improvement through learning from the data generated by itself. Note that the multi-contrast model can be also trained on data from fastMRI11, as shown in Figure 5. In summary, our proposed framework offers a general strategy for calibrationless parallel imaging reconstruction through learning from coil and protocol specific data. It is highly applicable to application scenarios where accurate coil sensitivity calibration is difficult.Acknowledgements
This work was supported in part by Hong
Kong Research Grant Council (R7003-19F, HKU17112120, HKU17127121 and
HKU17127022 to E.X.W., and HKU17103819, HKU17104020 and HKU17127021 to A.T.L.L.),
Lam Woo Foundation and Guangdong Key Technologies for Treatment of Brain
Disorders (2018B030332001) to E.X.W.
References
[1] Sodickson D. Parallel magnetic resonance imaging (or, scanners, cell phones, and the surprising guises of modern tomography). Med Phys 2007;34(6):2598-2598.
[2] Zhao B, Lu W, Hitchens TK, Lam F, Ho C, Liang ZP. Accelerated MR parameter mapping with low-rank and sparsity constraints. Magn Reson Med 2015;74(2):489-498.
[3] Uecker M, Lai P, Murphy MJ, Virtue P, Elad M, Pauly JM, Vasanawala SS, Lustig M. ESPIRiT-An Eigenvalue Approach to Autocalibrating Parallel MRI: Where SENSE Meets GRAPPA. Magn Reson Med 2014;71(3):990-1001.
[4] Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Med Image Comput Comput Assist Interv 2015;9351:234-241.
[5] Liu X, Pang Y, Jin R, Liu Y, Wang Z. Dual-Domain Reconstruction Networks with V-Net and K-Net for fast MRI. arXiv preprint arXiv:220305725 2022.
[6] Sriram A, Zbontar J, Murrell T, Zitnick CL, Defazio A, Sodickson DK. GrappaNet: Combining parallel imaging with deep learning for multi-coil MRI reconstruction. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2020, p 14315-14322.
[7] Souza R, Lucena O, Garrafa J, Gobbi D, Saluzzi M, Appenzeller S, Rittner L, Frayne R, Lotufo R. An open, multi-vendor, multi-field-strength brain MR dataset and analysis of publicly available skull stripping methods agreement. Neuroimage 2018;170:482-494.
[8] Chen C, Liu Y, Schniter P, Tong M, Zareba K, Simonetti O, Potter L, Ahmad R. OCMR (v1. 0)--Open-Access Multi-Coil k-Space Dataset for Cardiovascular Magnetic Resonance Imaging. arXiv preprint arXiv:200803410 2020.
[9] Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. arXiv preprint 2014;arXiv:1412.6980.
[10] Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L. Pytorch: An imperative style, high-performance deep learning library. Adv Neural Inf Process Syst 2019;32.
[11] Zbontar J, Knoll F, Sriram A, Murrell T, Huang Z, Muckley MJ, Defazio A, Stern R, Johnson P, Bruno M. fastMRI: An open dataset and benchmarks for accelerated MRI. arXiv preprint arXiv:181108839 2018.
Figures