4750
Multi-branch deep learning model integrated with multi-modal MRI for differential diagnosis of autism spectrum disorders
Xuan Yu1 and Meiyun Wang1,2
1Henan Provincial People’s Hospital & the People’s Hospital of Zhengzhou University, Zhengzhou, China, 2Laboratory of Brain Science and Brain-Like Intelligence Technology, Institute for Integrated Medical Science and Engineering, Henan Academy of Sciences, Zhengzhou, China
1Henan Provincial People’s Hospital & the People’s Hospital of Zhengzhou University, Zhengzhou, China, 2Laboratory of Brain Science and Brain-Like Intelligence Technology, Institute for Integrated Medical Science and Engineering, Henan Academy of Sciences, Zhengzhou, China
Synopsis
Keywords: Brain Connectivity, Brain Connectivity
This study proposes a multi-branch deep learning model fused with multi-modal MRI for the differential diagnosis of ASD. We solve the problem that traditional ASD classification algorithms based on static functional network connections ignore the time-varying characteristics of brain functional connections. Study the spatiotemporal characteristics of ASD brain imaging, and mine the information of functional connectivity between brain regions over time.Introduction/Purpose
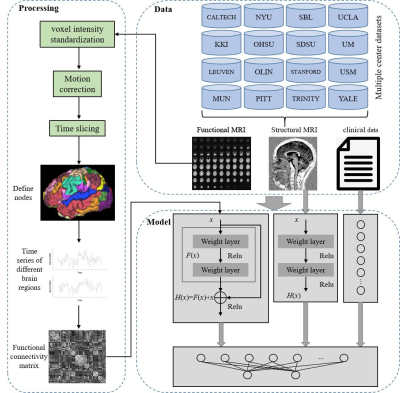
In order to effectively analyze the spatiotemporal data of autism spectrum disorders (ASD)[1] brain imaging, mine the information that the functional connections between brain regions change with time, and solve the problem that the traditional ASD classification algorithm based on static functional network connection ignores the time-varying characteristics of brain functional connections, a multi-branch deep learning model for ASD differential diagnosis fused with multi-modal magnetic resonance imaging (MRI) was proposed.Method
MRI and clinical data of 856 subjects (including 525 ASD subjects) were collected from ABIDE database. Firstly, the blood oxygen level dependent (BOLD) data of functional MRI are preprocessed by time slicing, motion correction, and voxel intensity standardization[2]. Then, based on CC200 atlas[3], the whole brain is divided into 200 regions of interest with different functions, from which a seed point is randomly selected for each brain region to obtain the corresponding BOLD signal sequence at different times. The sliding window strategy is used to segment 200 BOLD signal sequences to generate three-dimensional matrix data. Then, the method of transfer entropy in information entropy is applied to analyze the information transfer status between brain regions at different time scales, mining the temporal and spatial information of functional connectivity between brain regions at different time segments, and generating a three-dimensional dynamic brain functional connectivity matrix. Finally, a multi-modal fusion multi-branch deep learning algorithm is constructed to automatically mine the information in different modal data, and the residual convolution branch is applied to analyze the features in the three-dimensional dynamic brain function connection matrix. By analyzing T1 sequence attributes of structural MRI through deep convolution branching, the obtained multi-modal features are fused with clinical information. The differential diagnosis between ASD and normal developing subjects can be achieved by the full connection layer.Result
Through the transfer entropy method, we found that ASD subjects had the max information transfer value between the cingulate and the occipital pole region. Based on the information transfer status between different regions in functional MRI data, structural MRI data and clinical characteristics, the proposed multi-modal multi-branch fusion deep learning model achieved an average accuracy of 71.8%, an average sensitivity of 78.61% and a specificity of 65.48% in multiple center studies, respectively. The performance is better than the existing advanced ASD classification algorithms.Conclusion
Using the transfer entropy method to calculate the dynamic correlation of brain regions and mining its features through the multi-branch deep learning convolution algorithm can examine the activity characteristics of different brain regions or parts and their interrelationships. At the same time, fusing the multi-modal MRI deep learning model is of great significance to improve the accuracy and objectivity of ASD diagnosis results.Acknowledgements
No acknowledgement found.References
[1] American Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-5. 5th ed. American Psychiatric Association Publishing, Washington, D.C. 2013.
[2] Cameron C, Yassine B, Carlton C, et al. The Neuro Bureau Preprocessing Initiative: open sharing of preprocessed neuroimaging data and derivatives. In Neuroinformatics, Stockholm, Sweden. 2013.
[3] Craddock RC, James GA, Holtzheimer PE, et al. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp. 2012;33:1914-1928.
DOI: https://doi.org/10.58530/2023/4750