4747
Using machine learning to evaluate values of six diffusion models to predict the efficacy of neoadjuvant chemotherapy for esophageal cancer1Shanghai Key Laboratory of Magnetic Resonance, East China Normal University, Shanghai, China, 2Department of Radiology, Affiliated Cancer Hospital of Zhengzhou University & Henan Cancer Hospital, Zhengzhou, China, 3MR Scientific Marketing, Siemens Healthcare, Shanghai, China
Synopsis
Keywords: Cancer, Tumor
We assessed the performance of diffusion models for assessing response to neoadjuvant chemotherapy (NACT) using machine learning. Firstly, features were extracted from the region of interest on different parametric maps of different diffusion models for esophageal squamous cell carcinoma (ESCC) patients and changes of the parameters (Δ parameter) before and after NACT (pre-NACT and post-NACT) were calculated. Then different Δ-NACT models and pre-NACT models were built for using features from different diffusion models. The results demonstrated that diffusion models may be used to predict the efficacy of NACT in ESCC patients.Introduction
For patients with locally advanced esophageal cancer, neoadjuvant chemotherapy (NACT) and surgery are the current standard treatments[1], which can improve the prognosis [2]. However, the treatment response varies considerably [3]. Therefore, prediction of pathological responses is of great significance for personalized treatment. DWI plays an increasingly role in predicting response [4, 5]. Here, we assessed the performance of six diffusion models: the continuous-time random-walk (CTRW), diffusion kurtosis imaging (DKI), fractional order calculus (FROC), intravoxel incoherent motion IVIM, mono-exponential (MONO) and stretched exponential model (SEM), for assessing response to NACT in patients with esophageal squamous cell carcinoma (ESCC).Methods
DataEighty-six patients received two cycles of chemotherapy followed by surgery. All 86 patients performed MRI scanning within one week before NACT and 44/86 patients had a second scan within 7 days before surgery. The study was approved by the Ethics Committee of Henan Cancer Hospital, and all participants signed the informed consent. An experienced pathologist blinded to imaging data assessed pathological response according to Tumor Regression Grading (TRG) system as four scores: TRG 3 (poor response), TRG 2 (minimal response), TRG 1 (moderate response) and TRG 0 (complete response).
Preprocess
The tumors were segmented using ITK-SNAP (www.itksnap.org). Two radiologists with more than five years of experience in reading images and a radiologist with 15 years of experience in MRI diagnosis to select the optimal segmentation. The extracted features were calculated from segmented ROI, using locally developed scientific software MRstation (GE Healthcare, Shanghai, China). The calculations for the six models used DWI images with b=0, 50, 100, 150, 200, 400, 500, 600, 800, 1000, 1500, 2000 s/mm2. The changes of these parameters (Δ parameter) were calculated by subtracting pre-NACT value from and the corresponding post-NACT value.
Model construction
The flowchart of the model construction was illustrated in Figure 1.
Forty-four cases (17/27= response/non-response) were used to train the Δ-NACT (post-pre) model using leave-one-out cross validation (CV) approach. All features were normalized using z-score and for each pair of features whose Pearson correlation coefficient (PCC) larger than 0.99, one random feature in the pair was removed. Then, different combinations of three feature selectors (analysis of variance: ANOVA; Relief; recursive feature elimination: RFE) and two classifiers (support vector machine: SVM; logistic regression: LR) were tried out to find the best model. The number of features retained was determined by selecting the model with the best CV AUC. The number of features was limited to no more than 10.
Eighty-six cases (69/17=response/non-response) were used to train the pre-NACT model. The process of model building was same as mentioned above, except that a 5-fold CV was used instead of leave-one-out.
All the above processes were implemented using open-source FeatureExplorer [6].
Results
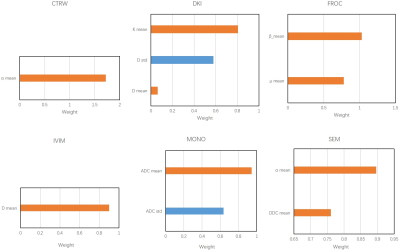
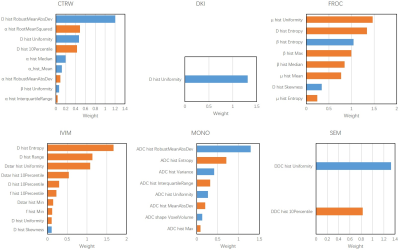
A total of 360 features were extracted from the 3D ROI of each patient, of which 330 radiomics features were used to build radiomics models and 30 quantitative features were used to build mean models. The combination of Relief and SVM was found to be best for Δ-NACT model and the pre-NACT model.According to the Table 1, the model with the best prediction response to NACT was the DKI model constructed by Δ_NACT features, and the CV AUCs were 0.575 (mean) and 0.871 (radiomics), which were much greater than the other five models: CTRW (0.869, 0.830), FROC (0.838, 0.739), IVIM (0.745, 0.783), MONO (0.685, 0.834), and SEM (0.791, 0.822). Features and their corresponding coefficients were shown in Figure 2 and Figure 3. The features contributed the most to the six models were: CTRW α mean, CTRW D histogram RobustMeanAbsDev, DKI K mean, DKI D histogram Uniformity, FROC β mean, FROC μ histogram Uniformity, IVIM D mean, IVIM D histogram Entropy, Mono ADC mean, Mono ADC histogram RobustMeanAbsDev, SEM α mean, and SEM DDC histogram Uniformity.
Discussion
In our study, the radiomics model in DKI constructed by Δ_NACT features showed the highest diagnostic value in predicting the efficacy of NACT in patients with locally advanced ESCC, and D Uniformity contributing the most to the model. This is similar to the findings of DKI in somatic tumour [7]. Furthermore, Song, et al [8] found that the IVIM ΔD value was a valid biomarker for predicting pathological response to NACT in patients with locally advanced ESCC, and Yu, et al [9] suggested that the K parameter in DKI has a higher diagnostic value as a representative indicator of the DKI model. So, the D parameter in DKI could likewise be a new biomarker for the efficacy of NACT in ESCC. The CTRW model can quantify voxels and their surroundings. Qin, et al [10] and Karaman, et al [11] concluded that the CTRW model has the potential to provide additional information on the prognosis and intrinsic subtype classification of breast cancer, and assess gliomas to complement histopathology. Our study is the first to apply the CTRW model to ESCC and confirms its value in reflecting the temporal and spatial heterogeneity of ESCC with delta features.Conclusion
Our study suggests that DWI diffusion models based on multiple b-values may predict the efficacy of NACT in ESCC patients.Acknowledgements
Long Cui and Bingmei Bai contributed equality to this study.References
1. Ajani J, D'Amico T, Bentrem D, et al. Esophageal and Esophagogastric Junction Cancers, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. Journal of the National Comprehensive Cancer Network : JNCCN. 2019;17(7):855-883.
2. Ando N, Kato H, Igaki H, et al. A randomized trial comparing postoperative adjuvant chemotherapy with cisplatin and 5-fluorouracil versus preoperative chemotherapy for localized advanced squamous cell carcinoma of the thoracic esophagus (JCOG9907). Annals of surgical oncology. 2012;19(1):68-74.
3. Yang H, Liu H, Chen Y, et al. Neoadjuvant Chemoradiotherapy Followed by Surgery Versus Surgery Alone for Locally Advanced Squamous Cell Carcinoma of the Esophagus (NEOCRTEC5010): A Phase III Multicenter, Randomized, Open-Label Clinical Trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2018;36(27):2796-2803.
4. Borggreve A, Goense L, van Rossum P, et al. Preoperative Prediction of Pathologic Response to Neoadjuvant Chemoradiotherapy in Patients With Esophageal Cancer Using F-FDG PET/CT and DW-MRI: A Prospective Multicenter Study. International journal of radiation oncology, biology, physics. 2020;106(5):998-1009.
5. Pellat A, Dohan A, Soyer P, Veziant J, Coriat R, Barret M. The Role of Magnetic Resonance Imaging in the Management of Esophageal Cancer. Cancers. 2022; 14(5).
6. Song Y, Zhang J, Zhang YD, et al.FeAture Explorer (FAE): A tool for developing and comparing radiomics models. PLoS One. 2020;15(8):e0237587.
7. Jia Fu, Lei Tang, Zi-Yu Li, et al.Diffusion kurtosis imaging in the pediction of poor reponses of locally advanced gastric cancer to neoadjuvant chemotherapy. European Journal of Radiology. 2020;128 (2020) 108974.
8. Song T, Yao Q, Qu J, Zhang H, et al. The value of intravoxel incoherent motion diffusion-weighted imaging in predicting the pathologic response to neoadjuvant chemotherapy in locally advanced esophageal squamous cell carcinoma. European Radiology. 2020; 31(3):1391-1400.
9. Jing Yu, Qing Xu, Jiacheng Song, et al. The value of diffusion kurtosis magnetic resonance imaging for assessing treatment response of neoadjuvant chemoradiotheraphy in locally advanced rectal cancer. Eur Radiol; DOI 10.1007/s00330-016-4529-6.
10. Yanjin Qin, Caili Tang, Qilan Hu, Jingru Yi, Ting Yin, Tao Ai. Assessment of Prognostic Factors and Molecular Subtypes of Breast Cancer With a Continuous-Time Random-Walk MR Diffusion Model: Using Whole Tumor Histogram Analysis. J.MAGN.RESON.IMAGING. 2022; DOI 10.1002/jmri.28474.
11. Karaman MM, Zhang J, Xie KL, Zhu W, Zhou XJ. Quartile histogram assessment of glioma malignancy using high b-value diffusion MRI with a continuous-time random-walk model. NMR Biomed. 2021;34(4):e4485.
Figures
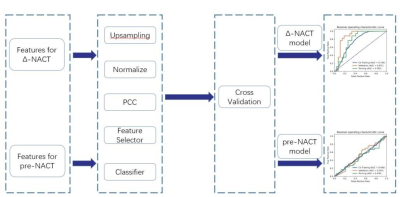
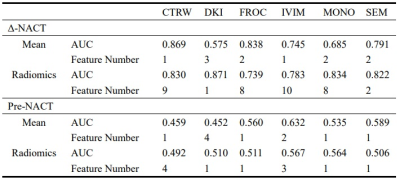
Table 1. The AUC and the number of features for the models.