4746
Improving Across-Dataset Schizophrenia Classification with Structural Brain MRI Using Multi-scale Transformer1Biomedical Engineering, Columbia University, New York, NY, United States, 2BME, Columbia University, New York, NY, United States, 3Department of Psychiatry, Columbia University, New York, NY, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Brain, Deep Learning
Schizophrenia is a neurological disorder that requires accurate and rapid detection for earlier intervention. Previous explorations in artificial intelligence showed overwhelming performance using deep learning in schizophrenia classification, though the generalization remained a challenge. We propose our 3D Multi-scale Transformer (MST) using T1W structural MRI data to detect schizophrenia. By synthesizing reconstructed images at different scales, the transformer-based architecture improves robustness to generalize in unseen data. The proposed method reaches the same-level performance of AUROC to the benchmark mark model in schizophrenia identification, and performs better in all leave-one-site-out generality tests.Introduction
Schizophrenia is a chronic progressive disorder that affects barely 1% of the population in the United States in 2022. This neuropsychiatric disease may result in disorganized behavior, delusions, and other serious symptoms that disable patients’ daily functioning. Studies have found its strong dependency on structural abnormalities in the brain such as ventricular enlargement[1]. Neuroimaging research has investigated brain alterations caused by schizophrenia using Magnetic Resonance Imaging (MRI). T1-weighted (T1W) MRI demonstrates differences in the T1 relaxation times of tissues and is widely applied to elucidate progressive changes in the schizophrenic brain [2]. Deep Learning is a computational technique that arose in the past 10 years to support the automated detection of Schizophrenia. While convolutional neural networks have been proven to outperform conventional machine learning models [3][4], the transformer architecture that emerged recently uses attention mechanisms to achieve even better results in medical image analysis and disease classification[5]. We design and propose our 3D Multi-scale Transformer (MST) inspired by the Vision Transformer (ViT) [6] which significantly enhances the across-datasets classification of schizophrenia.Methods
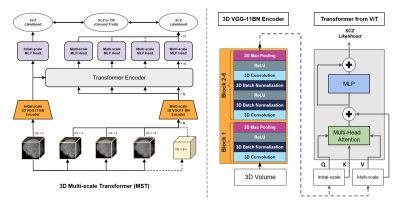
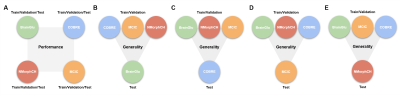
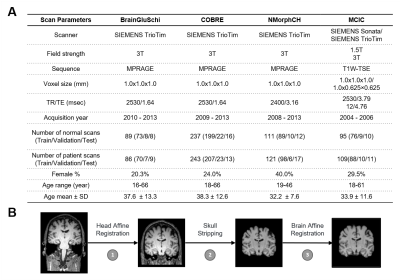
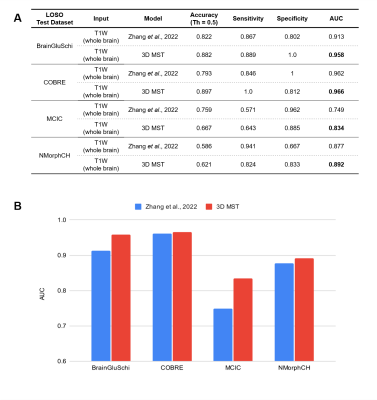
The architecture of our proposed 3D Multi-scale Transformer is shown in Figure 1. Two identical encoders extract features from the initial scale input and reconstructed multi-scale input. The encoders are 3D VGG-11BN backbones[7] with back five convolutional blocks. We downsample the input 3D T1W MRI images at a ratio of 2 as the initial scale and input to the first encoder. Then we downsample and upsample the initial images at a ratio of increasing powers of 2 ( ×2, ×4, ×8). The images keep the same size after this step but have a larger receptive field and more dilated information. The features are fed into a transformer encoder with spatial position embedding and batch normalization removed. To compute the attention, features are projected by 1×1 convolutional layers into three different spaces: ‘key’ and ‘value’; and ‘query’, separately. Lastly, linear voting at the decoding stage fuses the output to compute the loss. We evaluated model performance and generality using 4 datasets from the studies: BrainGluSchi [8], COBRE[9], NMorphCH[10], and MCICShare[11]. We tested the overall results using all available data and conducted a leave-one-site-out (LOSO) test to compare the generality. The across-datasets experiments are concluded in Figure 2. The T1-weighted structural MRIs in the 4 studies are gathered from the SchizConnect database (http://schizconnect.org/) and are summarized in Figure 3A. All datasets have homogeneous acquisition parameters and characteristics, except the MCICShare which has different MRI sequences and acquisition parameters. The whole-head T1W scans were registered to the MNI152 unbiased template by robust affine registration [12, 13]. We applied skull-stripping with the Brain Extraction Tool[14] before another affine registration of the whole-brain scans. The data preprocessing steps to remove unwanted artifacts and transform the data into a standard format are illustrated in Figure 3B. All datasets were merged and split into 8:1:1 train/validation/test groups for deep learning analysis. We used a learning rate of 10-5 and a CrossEntropy loss function. Each input 3D subject was cropped to (128×192×128) and output two scores of probability that correspond to the group label of schizophrenia patients or healthy controls, respectively.Results
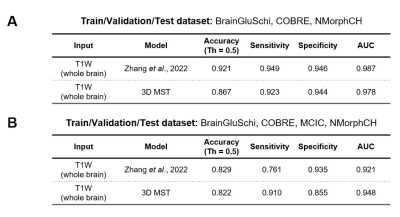
When training and testing on all 4 datasets, our 3D Multi-scale Transformer (MST) model performs better than the state-of-the-art model with an area under the receiver operating characteristic (AUC) of 0.948. When we excluded the MCICShare dataset, our proposed model almost replicated the best existing model performance. The results are compared in Figure 4. Our model outperformed the benchmark model in all leave-one-site-out generality tests, as shown in Figure 5. If training the model using COBRE, NMorphCH, and MCICShare datasets and testing on the BrainGluschi dataset, the baseline model performance shows a significant drop in terms of the accuracy, sensitivity, specificity, and AUC, while our proposed model remains robust towards the unseen dataset. This better generalization could also be revealed via another 3 tests in which our proposed model all beat the benchmark. Specifically, when testing on the MCICShare dataset alone, this effect becomes more significant.Discussion
Generalization is always a formidable challenge in deep learning tasks for MRI image analysis. Current models might have performed extremely well on the regular training-testing pipeline, but those numbers of performance, especially the specificity, unavoidably decreased when switching to an untrained target test. This gap makes the model no longer a potentially automatic diagnostic tool for clinical use, but instead, not convincing until trained with additional data. Our proposed model solves this problem by raising the idea of multi-scaling at the input stage. This method enables each fundamental element of the image to absorb useful information from adjacent ones and fuses the features to be more representative. By picking different numbers of the scaling factor, we are able to decide the limit of the receptive field and the number of images for each encoder. The attention mechanism in the transformer fuses extracted features and further enhanced the predictive ability of this deep-learning architecture. We use the same modality of 3D MRI data, with same-level computational cost, remain the model's robustness but significantly improve the generalizability. This approach could improve current MRI research in schizophrenia with various sites, sequences, and field strengths.Acknowledgements
This study was supported by the Zuckerman Mind Brain Behavior Institute at Columbia University and Columbia MR Research Center site.References
1. DeLisi, L.E., Szulc, K.U., Bertisch, H.C., Majcher, M. and Brown, K., 2022. Understanding structural brain changes in schizophrenia. Dialogues in clinical neuroscience.
2. Iwatani, J., Ishida, T., Donishi, T., Ukai, S., Shinosaki, K., Terada, M. and Kaneoke, Y., 2015. Use of T1‐weighted/T2‐weighted magnetic resonance ratio images to elucidate changes in the schizophrenic brain. Brain and behavior, 5(10), p.e00399.
3. Yassin, W., Nakatani, H., Zhu, Y. et al. Machine-learning classification using neuroimaging data in schizophrenia, autism, ultra-high risk and first-episode psychosis. Transl Psychiatry 10, 278 (2020).
4. Zhang, J., Rao, V.M., Tian, Y., Yang, Y., Acosta, N., Wan, Z., Lee, P.Y., Zhang, C., Kegeles, L.S., Small, S.A. and Guo, J., 2022. Detecting schizophrenia with 3d structural brain mri using deep learning. arXiv preprint arXiv:2206.12980.
5. Ashish V, Noam S, Niki P, Jakob U, Llion J, Aidan G, Lukasz K, and Illia P. Attention is all you need. ArXiv:1706.03762, 2017.
6. Dosovitskiy, A., Beyer, L., Kolesnikov, A., et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. ArXiv, abs/2010.11929.
7. Simonyan, K. and Zisserman, A., 2014. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556.
8. Mustafa C, Jon H, Barnaly R, et al. Multimodal classification of schizophrenia patients with MEG and fMRI data using static and dynamic connectivity measures. Front Neurosci, 10:466, oct 2016.
9. Alexandre S, Darya C, and Manuel G. Computer aided diagnosis of schizophrenia on resting state fMRI data by ensembles of ELM. Neural Networks: The Official Journal of the International Neural Network Society, 68:23–33, 2015.
10. Kathryn A, Todd P, Alexandr K, Daniel M, and Lei W. The northwestern university neuroimaging data archive (nunda). NeuroImage, 124:1131–1136, 2016.
11. Gollub, R.L., Shoemaker, J.M., King, M.D., White, T., Ehrlich, S., Sponheim, S.R., Clark, V.P., Turner, J.A., Mueller, B.A., Magnotta, V. and O’Leary, D., 2013. The MCIC collection: a shared repository of multi-modal, multi-site brain image data from a clinical investigation of schizophrenia. Neuroinformatics, 11(3), pp.367-388.
12. Mark J and Stephen S. A global optimisation method for robust affine registration of brain images. Medical Image Analysis, 5(2):143–156, 2001.
13. Mark J, Peter B, Michael B, and Stephen S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage, 17(2):825–841, 2002.
14. Stephen S. Fast robust automated brain extraction. Human Brain Mapping, 17(3):143–155, 2002.
Figures