4745
A Deep Learning Nomogram Based on Gd-EOB-DTPA MRI for Predicting Early Recurrence in Hepatocellular Carcinoma after Hepatectomy1Department of Radiology, The First Affiliated Hospital of Jinan University, Guangzhou, China, 2Neusoft Medical Systems Co., Ltd, Shanghai, China, 3Department of Radiology, Zhujiang Hospital of Southern Medical University, Guangzhou, China, 4Department of Liver Surgery, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Cancer
Prognostic risk assessment after hepatectomy for patients with hepatocellular carcinoma (HCC) remains difficult. Previous studies have shown that Gd-EOB-DTPA MRI is sensitive and accurate for HCC detection, but studies in predicting early recurrence after hepatectomy based on deep learning (DL) are still lacking. This study investigated the performance of a Gd-EOB-DTPA MRI-based DL approach, and then evaluated the DL nomogram incorporating deep features and significant clinical indicators. DL nomogram outperformed the clinical nomogram (validation AUC: 0.909 vs. 0.715). The proposed DL nomogram could provide a noninvasive and comprehensive tool for predicting early recurrence of HCC after curative resection.Introduction
Hepatic resection is the first-line treatment for patients with early-stage HCC and well-preserved liver function, but the recurrence rate of HCC is as high as 70% five years after surgery.1 The accurate prediction of post-hepatectomy early recurrence in patients with HCC is crucial for decision-making regarding postoperative adjuvant treatment. Numerous tumor factors have been identified as predictors of early recurrence, 2-5 however, most of the risk factors can only be obtained by postoperative pathology and cannot be used to assess prognosis and develop treatment plans prior to hepatectomy. Although several imaging features observed on MRI have been demonstrated to be associated with early recurrence, 6, 7 the prediction may be subjective and dependent on the radiologist's experience. Deep learning (DL) has been proved to have strong predictive value for the prediction of recurrence after liver transplantation and exploration of prognostic indicators in the HCC pathological images.8, 9 Studies using ethoxybenzyl diethylenetriamine pentaacetic acid (Gd-EOB-DTPA) MRI and DL methods to predict early recurrence after hepatectomy for HCC patients are still lacking. To demonstrate the capability of DL in predicting prognosis of HCC, we aimed to investigate the feasibility of deep features extracted from MRI with Gd-EOB-DTPA for predicting early recurrence after hepatic resection. Furthermore, we evaluated the predictive performance of the DL-based nomogram incorporating deep features and significant clinical indicators.Methods
A total of 285 HCC patients who underwent Gd-EOB-DTPA MRI scans within one month before curative resection were consecutively included. Clinical, pathological, and laboratory data were collected. The patients were randomly divided into the training and validation sets at a ratio of 7:3 (Table 1). Univariate logistic regression analysis was performed in the training set, and significant variables with P<0.05 were entered into the multivariate logistic regression using the forward likelihood ratio method to constructed the clinical nomogram. For the multi-sequence (mp) MRI acquisition, arterial phase (AP), portal venous phase (PVP), and hepatobiliary phase (HBP) images, were obtained for all the patients. Offset field correction and gray normalization were applied to the images to reduce the influence of scanning parameters, protocols, and individual differences. Two radiologists with 5 and 10 years of experience in diagnostic abdominal imaging independently delineated the tumor region without prior knowledge of the pathological findings, and when they disagreed on the results, the decision was reconciled. A state-of-the-art architecture VGGNet-19 pretrained on the ImageNet dataset was used as the DL feature extractor (Figure 1). The representative slice with the largest size of tumor region was duplicated into three channels (512*512*3) as the input, and then 1472 DL features were extracted from the AP, PVP, and HBP images, respectively. After selecting the features strongly related to early recurrence with feature value preconditioning, de-redundancy, and dimensionality reduction, different classifiers were investigated to establish a DL signature to identify the outcome status of early recurrence for each patient. The area under the curve (AUC), sensitivity, and specificity were used to evaluate the model performance while the nomogram, decision curve analysis (DCA) was conducted to confirm the clinical utility.Results
The performance comparisons among the models were provided in Table 2. Neutrophil count, AST, and MVI were identified as independent risk factors for early recurrence after hepatectomy and then used to build the clinical nomogram (Figure 2A), of which the training and validation AUCs were lower than both the single-layered and mp-MR-based DL signature (P<0.05*). Moreover, the performance of the mp-MR-based DL signature outperformed all the single-layered DL signatures. Furthermore, DL nomogram (Figure 2B) incorporating the tumor number (P=0.001*), microvascular invasion (P=0.039*), and mp-MR-based DL signature (P<0.001*) was the best for early recurrence prediction after hepatectomy, yielding AUCs of 0.949 and 0.909 in the training and validation sets, respectively. Excellent calibration curve and DCA confirmed the clinical usefulness of the DL nomogram (Figure 2C-F).Discussion
Several studies have explored the feasibility of clinic-radiological variables for predicting early recurrence of HCC, which is consistent with our findings.10-13 Previous radiomics studies have demonstrated the preponderance of radiomics analysis compared to clinical features.12, 13 More recent studies have shifted towards the field of deep learning. DL methods have outperformed the radiomics features for mass differentiation and prognosis of HCC in the latest research.14, 15 However, studies using Gd-EOB-DTPA MRI and DL methods to predict early recurrence after hepatectomy for HCC patients are still lacking. Our study is among the first that explored DL features extracted from Gd-EOB-DTPA MRI images to predict early recurrence after hepatectomy. Consist with the previous studies, 12-13 our results showed that the mp-MR-based signature offers more predictive power than the single-layered DL signatures. Furthermore, the nomograms by integrating the mp-MR-based signature and clinical-radiological factors provided multi-dimensional information, achieving higher predictive accuracy than the previous similar study.16 The proposed DL nomogram could provide a noninvasive and comprehensive preoperative tool for early recurrence risk assessment of hepatectomy, which is conducive to timely adjuvant therapy and monitoring.Acknowledgements
No acknowledgement found.References
1. Ishizawa T, Hasegawa K, Aoki T, et al. Neither multiple tumors nor portal hypertension are surgical contraindications for hepatocellular carcinoma. Gastroenterology. 2008;134(7):1908-1916.
2. Lee S, Kang T W, Song K D, et al. Effect of microvascular invasion risk on early recurrence of hepatocellular carcinoma after surgery and radiofrequency ablation. Ann Surg. 2021;273(3):564-571.
3. Hirokawa F, Hayashi M, Asakuma M, et al. Risk factors and patterns of early recurrence after curative hepatectomy for hepatocellular carcinoma. Surg Oncol. 2016;25(1):24-29.
4. Kwon S K, Yun S S, Kim H J, et al. The risk factors of early recurrence after hepatectomy in hepatocellular carcinoma. Ann Surg Treat Res. 2014;86(6):283-288
5. Li X, Qi Z, Du H, et al. Deep convolutional neural network for preoperative prediction of microvascular invasion and clinical outcomes in patients with HCCs. Eur Radiol. 2022;32(2):771-782.
6. Lee S, Kim S H, Lee J E, et al. Preoperative gadoxetic acid–enhanced MRI for predicting microvascular invasion in patients with single hepatocellular carcinoma. J Hepatol. 2017;67(3):526-534.
7. An C, Kim D W, Park Y N, et al. Single hepatocellular carcinoma: preoperative MR imaging to predict early recurrence after curative resection. Radiology. 2015;276(2):433-443.
8. Nam J Y, Lee J H, Bae J, et al. Novel model to predict HCC recurrence after liver transplantation obtained using deep learning: a multicenter study. Cancers. 2020;12(10):2791.
9. Shi J Y, Wang X, Ding G Y, et al. Exploring prognostic indicators in the pathological images of hepatocellular carcinoma based on deep learning. Gut. 2021;70(5):951-961.
10. Zhou L, Wang S B, Chen S G, et al. Prognostic Value of ALT, AST, and AAR in Hepatocellular Carcinoma with B-Type Hepatitis-Associated Cirrhosis after Radical Hepatectomy. Cli Lab. 2018;64(10):1739-1747.
11. Geh D, Leslie J, Rumney R, et al. Neutrophils as potential therapeutic targets in hepatocellular carcinoma. Nat Rev Gastro Hepat. 2022;19(4):257-273.
12. Zhao Y, Wu J, Zhang Q, et al. Radiomics analysis based on multiparametric MRI for predicting early recurrence in hepatocellular carcinoma after partial hepatectomy. J Magn Reson Imaging. 2021;53(4):1066-1079.
13. Chong H H, Yang L, Sheng R F, et al. Multi-scale and multi-parametric radiomics of gadoxetate disodium–enhanced MRI predicts microvascular invasion and outcome in patients with solitary hepatocellular carcinoma≤ 5 cm. Eur Radiol. 2021;31(7):4824-4838.
14. Yasaka K, Akai H, Abe O, et al. Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology. 2018;286(3):887-896.
15. Chaudhary K, Poirion O B, Lu L, et al. Deep Learning–Based Multi-Omics Integration Robustly Predicts Survival in Liver CancerUsing Deep Learning to Predict Liver Cancer Prognosis. Clin Cancer Res. 2018;24(6):1248-1259.
16. Zhang L, Xia W, Yan Z P, et al. Deep learning predicts overall survival of patients with unresectable hepatocellular carcinoma treated by transarterial chemoembolization plus sorafenib. Front Oncol. 2020;10:593292
Figures
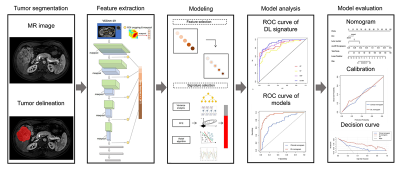
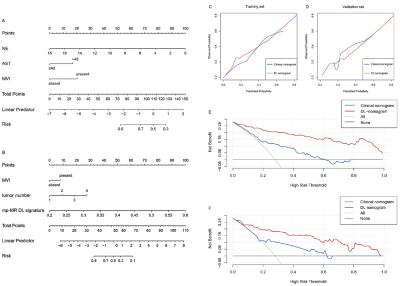
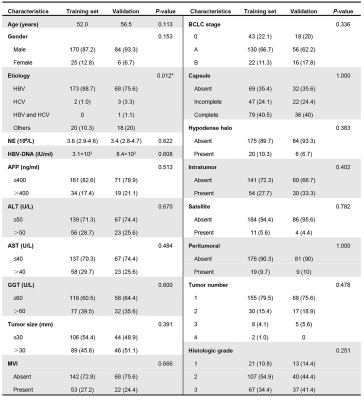
Table 1. Baseline characteristics of the training and validation sets
Note: Data are presented as number (%) or median (interquartile range, IQR); HBV, hepatitis B virus; HCV, hepatitis C virus; NE, neutrophil count; AFP, alpha-fetoprotein; ALT, alanine amino-transferase; AST, aspartate amino-transferase; GGT, glutamyltranspeptadase; BCLC, barcelona clinic liver cancer; AP, arterial phase; MVI, microvascular invasion.
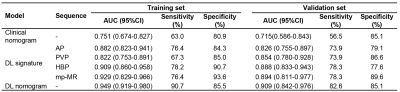
Table 2. Predictive performances of different models on the training and validation sets
Note: AUC, area under the curve; CI, confidence interval; mp, multi-sequence; DL, deep learning