4730
Reproducibility of 7T human cardiac 3D 31P-MRSI using concentric ring k-space trajectories (CRT).1Oxford Centre for Clinical MR Research (OCMR), RDM Cardiovascular Medicine, University of Oxford, Oxford, United Kingdom, 2Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 3Department of Imaging Methods, Institute of Measurement Science, Slovak Academy of Sciences, Bratislava, Slovakia
Synopsis
Keywords: Non-Proton, Heart, phosphorus
Cardiac 31P-MRS provides insight into metabolism of the failing heart. The commonly employed 3D-MRSI acquisitions are rather slow, even at 7T. Fast readout trajectories, such as concentric ring (CRT) have been recently suggested to substitute the slow Cartesian sampling (CSI), however their repeatability is yet unknown. Our preliminary data suggest comparable intra-session repeatability, but somewhat lower inter-session repeatability for mid septal voxels using 2.5 min CRT in comparison to 6.5 min CSI. Similar trends were observed for higher resolution CRT.Introduction
Phosphorus magnetic resonance spectroscopy (31P-MRS) can probe the energy metabolism of the human heart in vivo by measuring the phosphocreatine to adenosine triphosphate concentration ratio (PCr/ATP), an indicator of heart failure (1), or the potentially more sensitive chemical kinetics of the oxidative phosphorylation system. In both cases fast scanning is desirable, to either fit 31P-MRS into a clinical protocol, or to mitigate the multiple acquisitions required for the latter metrics.Employing fast MRSI readout trajectories, it could be possible to leverage the full 2.8 times increase in SNR(2) to achieve close to the theoretical 7.8-times (2.82) speed increase when moving from 3 to 7 T. This is not possible using the most common approach, point-by-point Cartesian sampling (CSI). A new method employing a fast MRSI readout trajectory, concentric ring trajectory (CRT), has recently been described (3).
CRT can measure PCr/ATP maps as CSI sampling in a fraction of the time (with matched resolution) or with higher spatial resolution (in a matched time). While the repeatability of cardiac 3D 31P-MRSI sequences using Cartesian sampling at 7T is known (4), the repeatability of the CRT sequences is yet to be determined. Hence, the aim of this study was to evaluate the intra- and inter-session repeatability of three different CRT protocols.
Methods
Four sequences were evaluated: Cartesian acquisition-weighted MRSI, termed CSI henceforth, (matrix size 10x10x10, 4 averages at k=0, 6:31 mins) and the CRT sequence with a matched matrix size employing 12 (2:31 mins) and 10 rings (1:37 mins), and with a matrix size of 12x12x12 and 19 rings (6:55 mins, labelled as high-res CRT [HRCRT]). These four sequences were run twice in a session and on two different days, 72 hours apart, to evaluate intra- and inter-session repeatability (Figure 1 ).Three healthy, lean, young volunteers (1 female) were scanned supine in a Siemens Magnetom 7T scanner (Siemens, Erlangen) equipped with a square surface transmit and 16 channel receive array coil (Rapid Biomedical) positioned over the heart (4). CSI was run with 240x240x200 mm3 FoV, 1 s TR, 8 kHz bandwidth, and 2048 time-samples. CRT had matched FoV and TR, but 2778 Hz bandwidth with 720 time-samples.
CSI data were reconstructed online and CONCEPT data offline using the non-uniform FFT (NUFFT) toolbox with min-max Kaiser-Bessel kernel interpolation and twofold oversampling (5) in MATLAB (MathWorks, Natick, USA). No density compensation was required. Individual coil data was combined using the WSVD algorithm (6). CSI/MRSI data fitting was performed using the OXSA toolbox (7).
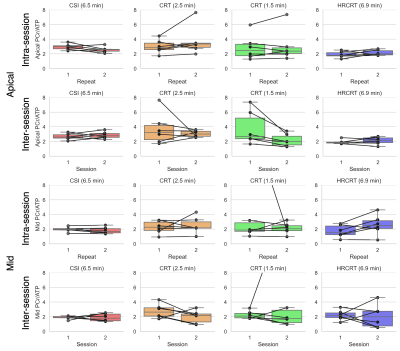
Sequences were compared using PCr/γ-ATP ratios, corrected for partial saturation and blood contamination, in mid-septal voxels from apical, mid, and basal slices of the heart. All corrections were calculated per-subject and per-session (2).
The intra-session variability was assessed through the mean and difference between PCr/ATP ratios from equivalent datasets within the same session, e.g., by comparison of datasets A1 with E1 and A2 with E2 of each volunteer for the CSI acquisitions. And the inter-session variability was assessed through the mean and difference between PCr/ATP ratios from equivalent datasets in both protocols for each subject, e.g, by comparing Dataset A1 with A2, and E1 with E2 in case of CSI (see Figure 1 for detailed description).
The coefficient of repeatability (CoR) was calculated from SD of the signed differences in PCr/ATP between two scans for each subject according to CoR = SDintrasubject × $$$\sqrt2$$$ × 1.96. A lower CoR reflects higher repeatability.
Results & Discussion
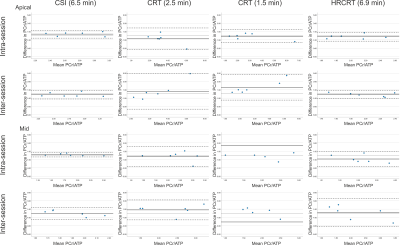
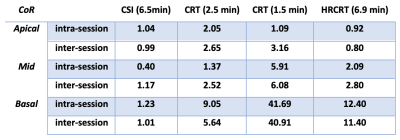
Figure 2 depicts the typical position of the analysed voxels as well as representative spectra from the mid septal voxel of one subject demonstrating good agreement between the acquired data for all sequences. There were no significant differences (paired t-test) in the calculated cardiac PCr/ATP between repeated measurements for all voxels, sequences, and repetitions (Figure 3). The Bland-Altman plots showing the repeatability of all sequences in apical and mid septal voxels are depicted in Figure 4.High repeatability was observed for the long CSI scan with CoR for mid septal voxel 0.40 for intra-session and 1.17 for inter-session comparison. While our subject numbers are currently low, this falls into the same range as previously reported repeatability of 8x16x8 CSI matrix by Ellis et al. with CoR of 0.67 and 0.75 for intra- and inter-session comparisons, respectively. To achieve the increase in speed, the CRT protocols sacrifice some signal-to-noise (SNR), which leads to higher CoR, i.e., 1.37 for the intra-session and 2.52 for the inter-session comparison of mid septal voxel of the 2.5 minute CRT sequence. For full report of the observed CoRs see Table 1. The high CoR values for basal voxels reflect the very low SNR suggesting that with the current setup these voxels are effectively not usable. This could be potentially mitigated by the use of a whole-body transmit coil (8).
Conclusion
Our preliminary results suggest comparable intra-session repeatability, but lower inter-session repeatability, of highly accelerated 3D-MRSI acquisition using CRT trajectory, providing either improved speed (2.5 minutes in comparison to 6.5 minutes CSI) and increased spatial resolution (6.66 mL in comparison to 11.5 mL nominal resolution).Acknowledgements
WTC acknowledges grant funding from the Wellcome Trust [225924/Z/22/Z] and [224430/Z/21/Z]. The Wellcome Centre for Integrative Neuroimaging is supported by core funding from the Wellcome Trust (203139/Z/16/Z and 203139/A/16/Z). LV is supported by a Sir Henry Dale Fellowship of the Wellcome Trust and the Royal Society [221805/Z/20/Z] and would also like to acknowledge the support of the Slovak Grant Agencies VEGA [2/0003/20] and APVV [19–0032].
References
1. Neubauer S. The Failing Heart — An Engine Out of Fuel. N Engl J Med 2007;356:1140–1151 doi: 10.1056/NEJMra063052.
2. Rodgers CT, Clarke WT, Snyder C, Vaughan JT, Neubauer S, Robson MD. Human cardiac 31P magnetic resonance spectroscopy at 7 tesla. Magnetic Resonance in Medicine 2014;72:304–315 doi: 10.1002/mrm.24922.
3. Clarke WT, Hingerl L, Strasser B, Bogner W, Valkovič L, Rodgers CT. Three-dimensional, 2.5-minute, 7T phosphorus magnetic resonance spectroscopic imaging of the human heart using concentric rings. NMR in Biomedicine n/a:e4813 doi: 10.1002/nbm.4813.
4. Ellis J, Valkovič L, Purvis LAB, Clarke WT, Rodgers CT. Reproducibility of human cardiac phosphorus MRS (31P-MRS) at 7 T. NMR in Biomedicine 2019;32:e4095 doi: 10.1002/nbm.4095.
5. Fessler JA, Sutton BP. Nonuniform fast Fourier transforms using min-max interpolation. IEEE Transactions on Signal Processing 2003;51:560–574 doi: 10.1109/TSP.2002.807005.
6. Rodgers CT, Robson MD. Receive array magnetic resonance spectroscopy: Whitened singular value decomposition (WSVD) gives optimal Bayesian solution. Magnetic Resonance in Medicine 2010;63:881–891 doi: 10.1002/mrm.22230.
7. Purvis LAB, Clarke WT, Biasiolli L, Valkovič L, Robson MD, Rodgers CT. OXSA: An open-source magnetic resonance spectroscopy analysis toolbox in MATLAB. PLoS One 2017;12:e0185356 doi: 10.1371/journal.pone.0185356.
8. Valkovič L, Dragonu I, Almujayyaz S, et al. Using a whole-body 31P birdcage transmit coil and 16-element receive array for human cardiac metabolic imaging at 7T. PLOS ONE 2017;12:e0187153 doi: 10.1371/journal.pone.0187153.
Figures
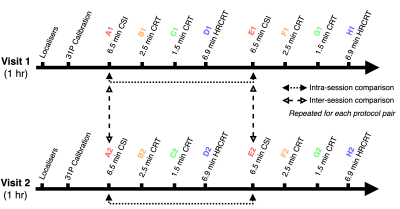
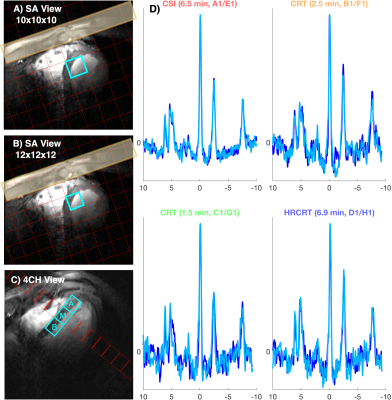
Figure 2: Signal localisation and example spectra. A & B show the lower resolution (10x10x10) and high-resolution (12x12x12) MRSI grid for overlaid on short axis localisers. Saturation bands suppress skeletal muscle signal (yellow patches). Voxels for analysis were selected from the mid intraventricular septum in an apical, mid, and basal slice of the heart (C). D Example spectra from mid-septal voxels of a single subject. The spectra shown are the repeated measurements within a single session.