4722
Accelerated in-vivo 23Na Multi-Quantum Coherences MRI by utilizing Low-Rank Matrix Completion1Computer Assisted Clinical Medicine, Heidelberg University, Mannheim, Germany, 2CRMBM, Aix-Marseille Université, Marseille, France
Synopsis
Keywords: Non-Proton, Multi-Contrast, Sodium
Sodium (23Na) MRI has received increased attention as a potential biomarker for disease states thanks to the advent of ultra-high field MRI. One interesting asset from 23Na MRI is the distinction between single and triple quantum signal to further characterize tissues. However, 3D 23Na multi-quantum coherences (MQC) imaging requires multiple radiofrequency phase-cycling, which is inherently time-consuming and therefore difficult to include in protocols. In this work, we propose to accelerate 23Na MQC MRI by leveraging multi-dimensional under-sampling coupled with a dedicated Low-Rank matrix completion image reconstruction.Introduction
Conventional sodium (23Na) MRI is a promising tool to probe tissue ionic homeostasis, but often remains limited to the analysis of the sodium signal intensity1. Thanks to its 3/2 spin, 23Na multi-quantum coherences MRI on the other hand enables to disentangle the underlying multi-quantum coherences (MQC) and therefore holds potentially richer sodium tissue characterization2 when compared to conventional 23Na signal. Especially the triple quantum (TQ) signal has shown to be sensitive to the intracellular sodium compartment and the molecular environment surrounding sodium atoms3. Unfortunately, 23Na MQC MRI requires a minimum of 12 RF phase cycling steps, which prolongs acquisition time up to one hour and is thus impractical for large scale in-vivo studies. Alternatively, 23Na MQC MRI exhibits multi-dimensional data, which is therefore intrinsically redundant and can be exploited to reconstruct undersampled 23Na MQC MRI data. Thus, for the first time we propose to utilize low-rank matrix completion to reliably reconstruct phase-cycled under-sampled phantom, in-vivo knee and brain 23Na MQC MRI.Methods
All measurements were performed on 3T MRI (Trio, Siemens Heathineers, Erlangen,Germany) with a bird-cage dual-tuned 23Na / 1H head coil (RapidBiomedical, Rimpar, Germany). 23Na MQC MR images were obtained by utilizing the CRISTINA sequence4, with the following parameters.In phantom: FoV 260x260x240mm3, matrix size 32x32x12, $$$\tau$$$ =8.1ms, BW=200Hz/px,TE/$$$\Delta$$$TE/NTE= 1.74/6.4ms/10, TR=140ms, 7 averages resulting in TA=2x30 minutes.
For knee in-vivo, one volunteer (42, m), FoV 270x270x200mm3,matrix size 26x26x10, $$$\tau$$$=5.7ms, BW=220Hz/px, TE/ $$$\Delta$$$TE/NTE=1.71/6.2ms/10, TR=150ms, TA=2x31.
For brain in-vivo, in three volunteers (28+/-2, f): FoV 230x230x160mm, matrix size 24x24x8, $$$\tau$$$=12.9ms, BW=210Hz/px, TE/$$$\Delta$$$TE/NTE= 1.62/6.2ms/10, TR=150ms resulting in TA=2x31min.
Image reconstruction: 23Na MQC raw data were processed either fully-sampled or with a retrospective variable density 3D Poisson-disk undersampling5 to simulate various acceleration rates. Sampling pattern was varied along the phase-cycle dimension to create complementary patterns. Finally, the images were reconstructed utilizing the SAKE6 framework, replacing the coil dimension with the phase-cycled dimension, and solving the following optimization problem:
$$ \min_u || \Phi_F(u) - f ||^2_2 < \sigma^2 s.t. u = H^*(A), rank(A) = k' $$
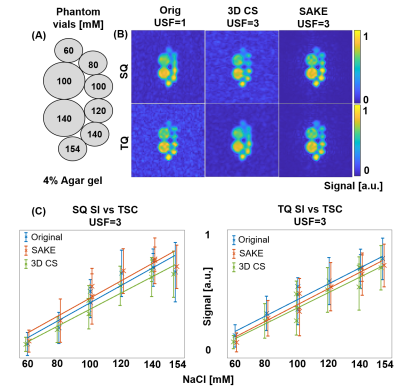
with $$$u$$$ being the image to reconstruct, $$$\Phi_F$$$ being the Fourier transform operator, $$$f$$$ the acquired data and $$$\sigma$$$ the noise signal variance . $$$k'$$$ is a prior estimate of $$$rank(A)$$$ of matrix $$$A$$$, enforcing subspace SVD and $$$H^*$$$ being the inverse structured Hankel matrix operator6. By reshaping multi-echo and phase-cycle dimensions, rank deficiency of the data matrix is thus enforced. Reconstruction performance was evaluated on phantom and in-vivo brain data by structural similarity (SSIM), root mean squared error (RMSE) and sodium quantification.
Results
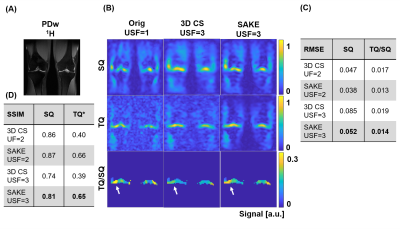
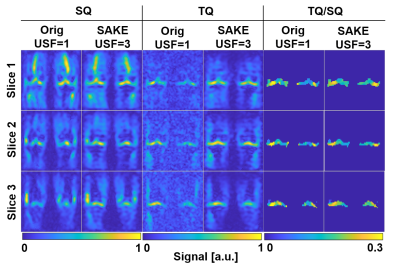
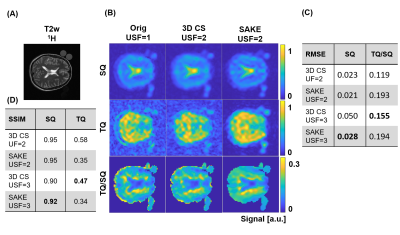
For 3-fold undersampled in-vivo knee 23Na MQC MRI, SAKE improved SSIM for the SQ image significantly by 0.07, when compared to 3D CS(Fig.3). Especially for higher acceleration factors, SAKE outperformed compressed sensing by reducing RMSE in SQ and TQ/SQ ratio. In addition, SAKE retained anatomical structures in the TQ images, due to shared information in 23Na MQC's multi-dimensional space, which resulted in degraded RMSE and SSIM. These findings were spatially consistent across several slices (Fig.4). SAKE images exhibited superior reconstruction performance on in-vivo brain data by reducing the RMSE for the SQ images by a factor of 2 but not for TQ images when compared to 3D CS( Fig.5).Discussion & Conclusion
Conclusively, we have demonstrated that low-rank matrix completion is an interesting framework to accelerate 23Na MQC MRI that can be used to reconstruct 3-fold undersampled in-vivo knee data. SAKE is especially well-suited to reconstruct in-vivo brain and knee SQ images, but fails at reconstructing TQ brain signals. This issue is attributed to shared information along the dimensions and the exploitation of subspace singular value decomposition. For instance, SQ signal exhibits relatively high signal amplitude when compared to TQ. Hence, SQ signal is coherent along temporal dimension, potentially biasing TQ image reconstruction. Further integration combining low-rank and MQC signal modeling as well as prospective undersampling to increase resolution will be investigated in the future to fully demonstrate the potential of low-rank matrix completion for 23Na MQC MRI.Acknowledgements
The author received financial support for the research by ISMRM research exchange grant program and PROCOPE Mobility 2022.References
1. Huhn K, Engelhorn T, Linker RA, Nagel AM. Potential of Sodium MRI as a Biomarker for Neurodegeneration and Neuroinflammation in Multiple Sclerosis. Front Neurol. 2019 Feb.
2. Hoesl, M.A., Kleimaier, D., Hu, R., Malzacher, M., Nies, C., Gottwald, E. and Schad, L.R. (2019), 23Na Triple-quantum signal of in vitro human liver cells, liposomes, and nanoparticles: Cell viability assessment vs. separation of intra- and extracellular signal. J. Magn. Reson. Imaging, 50: 435-444.
3. LaVerde, G., Nemoto, E., Jungreis, C., Tanase, C. and Boada, F. (2007), Serial triple quantum sodium MRI during non-human primate focal brain ischemia. Magn. Reson. Med., 57: 201-205.
4. Hoesl, M.A.U., Schad, L.R., Rapacchi, S.,Volumetric 23Na Single and Triple-Quantum Imaging at 7T: 3D-CRISTINA, Zeitschrift für Medizinische Physik, Volume 32, Issue 2, 2022, Pages 199-208.
5. Kustner, T., Schwartz, M., Würslin, C., Martirosian, P., Schwenzer, N., Yang, B., & Schmidt, H. (2016). Compressed Sensing LAB: An MR acquisition and reconstruction system. In Proceedings of the ISMRM Workshop on Data Sampling and Reconstruction.
6. Shin PJ, Larson PE, Ohliger MA, Elad M, Pauly JM, Vigneron DB, Lustig M. Calibrationless parallel imaging reconstruction based on structured low-rank matrix completion. Magn Reson Med. 2014 Oct;72(4):959-70.Figures