4693
Feasibility of Strain Encoded (SENC) MRI to Assess Cardiac Contractility on a Commercial 0.55T System1Ohio State University, Columbus, OH, United States, 2Myocardial Solutions, Inc., Morrisville, NC, United States
Synopsis
Keywords: Cardiomyopathy, Low-Field MRI, Strain
In phantom, human volunteers, and an animal model, this study shows the feasibility of strain encoding (SENC) on a commercially available 0.55T low field scanner with limited gradient performance. Measured through-slice strain in a deformation phantom agrees with steady-state free precession (SSFP) cine; global strains in healthy humans fall into the normal expected range; infarcted myocardial segments in a porcine model demonstrated reduced strain.INTRODUCTION
Low field CMR has gained recent attention as low field systems can provide access to a wider patient population due to lower manufacture, installation, and maintenance costs, reduced susceptibility artifact, and larger patient bore [1, 2]. Quantification of myocardial strain is also gaining acceptance for early detection of cardiac dysfunction, prior to overt functional or structural changes of the heart [3]. Strain Encoded MRI (SENC) is a reproducible method of directly measuring longitudinal and circumferential strain [4]. While low field scanners have shown promise for other cardiac MRI applications [1,2,5], the limited signal noise ratio imposes a challenge to SENC, and therefore we sought to investigate the performance of SENC at low field.METHODS
A prototype SENC sequence was implemented using a segmented k-space, spoiled gradient echo readout on a 0.55T system with 80 cm patient bore and maximum gradient amplitude 26mT/m and slew rate 45mT/m/ms (MAGNETOM Free.Max, Siemens Healthcare, Erlangen, Germany). The sequence was validated using a dynamic phantom that provides mechanical deformation of a gel block. Two settings were used to generate peak strains of approximately -20% and -30%. Healthy volunteers were scanned using a 6-channel flexible body array and 6 spine coil elements. An MRI compatible patient monitor was used for ECG triggering. Scan parameters were as follows: TE/TR 2.8ms/6.9ms, temporal resolution 27.5ms, FOV 380mm x 240mm, acquisition window 600ms, matrix 96x60, slice thickness 15mm, flip angle 10 deg, BW 200Hz/pixel, segments 4, acceleration rate 2, breath-hold duration 8 heartbeats. Three short-axis slices were used to measure global longitudinal strain (GLS), and 3 long-axis views to measure global circumferential strain (GCS) in right (RV) and left ventricles (LV). A 90 min coronary artery balloon occlusion followed by reperfusion was used to induce myocardial infarction in a porcine model. SENC images were obtained in 3 long axis and 3 short axis views to assess segmental longitudinal and circumferential strain in the left ventricle. Late Gadolinium Enhancement (LGE) images were obtained approximately 10 minutes after contrast injection to visually delineate infarcted segments. All images from phantom, human subjects, and animals were quantified in MyoStrain version 5.4 (Morrisville, NC USA, 27560). In the phantom, SENC results were compared to direct measurement of phantom deformation in cine images. In the human subjects, GLS and GCS were compared to published normal values [6]. In the animals, segmental strain was compared to the infarct region based the occluded artery, and on visual assessment of LGE images.RESULTS
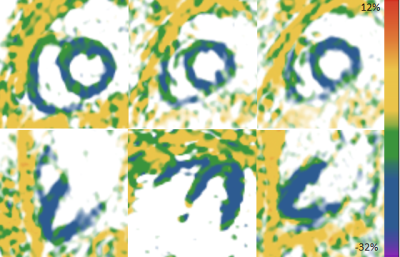
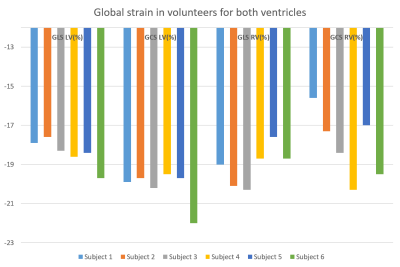
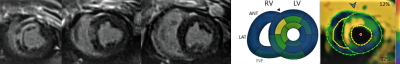
Strain values measured at phantom settings of -20% and -30% using cine vs. SENC were -20.4% vs -21.8%, and -28.4% vs -27.2%, respectively. Six healthy volunteers (5 male, mean age 46.3 ± 13.5 years) were scanned. Color is indicative of strain in the frames shown in Figure 1. Mean global strain values across volunteers (Figure 2) were (mean ± SD): GLS LV, -18.4% ± 0.7%; GCS LV, -20.2% ± 0.9%; GLS RV, -19.1% ± 1.0%; GCS RV, 18.0% ± 1.7%. These strain values are within the expected normal range for healthy hearts based on SENC data acquired at higher field [6]. Three Yorkshire pigs (31.5 ± 5.4 kg) were scanned 6 to 7 days after myocardial infarction. Myocardial segments within the infarct region confirmed by gadolinium enhancement demonstrated reduced strain by SENC in all animals (Figure 3).DISCUSSION
Deformation phantom results indicated good agreement with direct measurement, global strain values in volunteers fell within the expected normal range for healthy hearts, and strain deficits were detected in regions affected by myocardial infarction in an animal model. These results support the feasibility of SENC-based strain measurement at low field, despite the reduced signal and gradient performance of this scanner. The implementation used in this study was a segmented Cartesian k-space sampling mode requiring breath holding. Real-time, single-heartbeat SENC has been successfully used at higher field based on echo–planar and spiral k-space trajectories; these methods have yet to be explored at low field and may be challenged by the limited gradient performance of this particular system. This study included only a small number of healthy subjects and involved a single disease model at one site.CONCLUSION
This study demonstrates the feasibility of measuring global LV and RV strain, as well as regional strain deficits, with SENC on a commercially available, 0.55T scanner. Future studies are warranted in patients with cardiovascular disease.Acknowledgements
No acknowledgement found.References
1. Simonetti, Orlando P., and Rizwan Ahmad. "Low-field cardiac magnetic resonance imaging: a compelling case for cardiac magnetic resonance’s future." Circulation: Cardiovascular Imaging 10, no. 6 (2017): e005446.
2. Campbell-Washburn, Adrienne E., et al. "Opportunities in interventional and diagnostic imaging by using high-performance low-field-strength MRI." Radiology 293.2 (2019): 384.
3. Erley J, Zieschang V, Lapinskas T, Demir A, Wiesemann S, Haass M, Osman NF, Simonetti OP, Liu Y, Patel AR, Mor-Avi V. "A multi-vendor, multi-center study on reproducibility and comparability of fast strain-encoded cardiovascular magnetic resonance imaging." The international journal of cardiovascular imaging. 2020 May;36(5):899-911.
4. Osman, Nael F., et al. "Imaging longitudinal cardiac strain on short‐axis images using strain‐encoded MRI." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 46.2 (2001): 324-334.
5. Varghese J, Craft J, Crabtree CD, Liu Y, Jin N, Chow K, Ahmad R, Simonetti OP. Assessment of cardiac function, blood flow and myocardial tissue relaxation parameters at 0.35 T. NMR Biomed. 2020 Jul;33(7):e4317. doi: 10.1002/nbm.4317. Epub 2020 May 4. PMID: 32363644.
6. Korosoglou, Grigorios, et al. "Strain‐encoded magnetic resonance: a method for the assessment of myocardial deformation." ESC heart failure 6.4 (2019): 584-602.
Figures