4651
Accelerated Four-Dimensional Rosette J-resolved Spectroscopic Imaging (4D ROSE-JRESI) with semi-LASER Localization: A pilot study1Radiological Sciences, University of California, Los Angeles, Los Angeles, CA, United States, 2School of Health Sciences, Purdue University, West Lafayette, IN, United States, 3Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN, United States, 4School of Nursing and Brain Research Institute, University of California, Los Angeles, Los Angeles, CA, United States
Synopsis
Keywords: Data Acquisition, Brain
Undersampling spatial and spectral dimensions is essential to achieve clinically feasible scan times in multi-dimensional spectroscopic imaging. Sampling pattern of rosette trajectory has higher incoherence than that of the other non-Cartesian trajectories like spiral and radial, and can achieve higher compressed-sensing reconstruction performance. While rosette spectroscopic imaging has been attempted for 2D (2 spatial+1 spectral) and 3D (3 spatial+1 spectral) spectroscopic imaging, it has thus far not been shown for J-resolved spectroscopic imaging. In this pilot study, we implemented a rosette echo-planar J-resolved spectroscopic imaging (ROSE-JRESI) sequence and studied the feasibility of high acceleration factors for clinically feasible runtimes.Introduction
MR spectroscopy (MRS) is an efficient biochemical tool for non-invasively analyzing metabolite and lipid concentrations in human tissues (1-4). Compared to one dimensional spectrum, two-dimensional spectral variant resolves peak information along an additional spectral dimension which helps to disperse the spectrum better (1-4). However, acquisition of MRSI after adding the 2nd spectral encoding can increase the total acquisition time significantly. Even though k-space-weighted and average-weighted schemes have been used to shorten the total duration of MRSI, echo-planar spectroscopic imaging (EPSI) showed further acceleration of the total acquisition duration (5,6). Undersampling the spatial and 2nd spectral dimension is essential to achieve clinically feasible scan times. Compressed sensing (CS) based reconstruction techniques are known to be capable of recovering the signal depending on the signal sparsity and incoherent sampling patterns (7). However, non-cartesian J-resolved spectroscopic imaging has not been attempted so far. It has been reported that the sampling pattern of the rosette trajectory has higher incoherence than that of the other non-Cartesian trajectories like spiral and radial, and can thus achieve higher CS reconstruction performance (8). While rosette spectroscopic imaging has been attempted for 2D (2 spatial+1 spectral) and 3D (3 spatial+1 spectral) spectroscopic imaging (9-10), it has thus far not been shown for J-resolved spectroscopic imaging. In this pilot study, we implemented a four-dimensional (4D) rosette echo-planar J-resolved spectroscopic imaging (4D ROSE-JRESI) sequence and studied the feasibility of high acceleration factors for clinically feasible runtimes.Materials and Methods
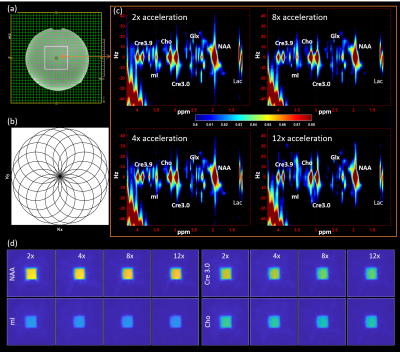
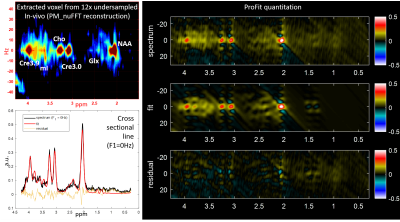
The 4D ROSE-JRESI sequence was designed based on the rosette trajectory for two spatial dimensions as described in (11) and was implemented on a Siemens 3T clinical scanner. Data from a brain phantom containing metabolites at physiological concentrations was acquired. A 68-year-old healthy volunteer was recruited with IRB approval for the acquisition of in vivo brain data. Phantom scans were acquired with a 32×32 matrix size, a 24×24×2cm3 slab using volumetric semi-LASER localization, TE=28.6ms, TR=1.5s, and a spectral width of 1250 Hz, spatially interleaved 16 petals, with 512 t2 points, 64 t1 points and 2 averages. It has been reported that number of petals = number of pixels in one dimension of reconstructed image is sufficient for adequate reconstruction (12). The phantom data were retrospectively undersampled at 4x (4x spatial (8 petals)), 8x (4x spatial and 2x t1 (32 t1 points)) and 12x (4x spatial and 3x t1 (22 t1 points)) to study the effect of higher undersampling. The volunteer scan was acquired with 8 petals and 22 non-uniformly sampled t1 points with decaying exponential sampling density, for a total of approx. 12x acceleration. All other parameters were kept the same as mentioned earlier. Time for one average was 4 minutes, 30 seconds including 4 dummy preparation scans and 8 minutes, 54 seconds for 2 averages. CS reconstruction using Perona-Malik (PM) (13-16) and non-uniform FFT (nuFFT) (17) was used to estimate the missing samples of k-space. ProFit quantitation was used to quantify the resulting spectra (18).Results
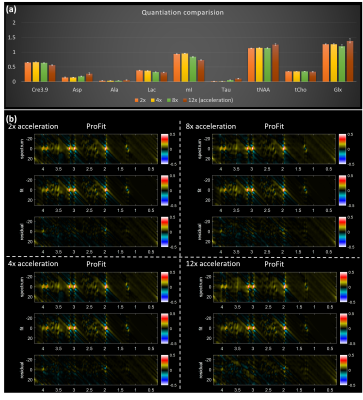
Figure 1 shows the localization image, rosette trajectory with 16 petals, extracted voxel from reconstructed data at 2x, 4x, 8x and 12x acceleration factors and different metabolite maps at multiple acceleration factors. The 2D spectrum, fit and residual based on ProFit quantitation for the voxels in Fig. 1(c) are shown in Fig. 2 along with a bar chart showing comparison of metabolite ratios with respect to Creatine 3.0 (Cre3.0) across multiple voxels from different acceleration factors. In-vivo data was reconstructed using nuFFT based PM. Sagittal and axial localization images, as well as the reconstructed metabolite maps of Cre 3.0, Creatine 3.9 (Cre 3.9), myo-Inositol (mI), N-acetylaspartate (NAA), choline (Cho) and Glx (Glutamine + Glutamate) overlaid on the localization image are shown in Figure 3. A multi-voxel spectrum covering all the voxels within the VOI (white box in the localization image) is shown in Figure 4. An extracted spectrum from the reconstructed in-vivo data and its ProFit quantitation are shown in Figure 5.Discussion
Rosette based J-resolved 4D spectroscopic imaging was implemented and tested using phantom and in-vivo datasets. The metabolite maps in Fig 1 shows reconstructed maps devoid without visible undersampling artifacts. Visible artifacts outside VOI was observed when only 8 petals were acquired corresponding to an 8x acceleration along kx and ky. It was observed that a better approach for 8x acceleration is 4x along spatial and 2x along t1 dimensions for reconstruction without any visible artefacts. Increasing acceleration beyond 12x tend to introduce more artifacts, most of the metabolite ratios stayed within relatively same range across different acceleration factors until 12x. Total NAA (tNAA), aspartate (Asp) and alanine (Ala) ratios where slightly overestimated while while mI, lactate (Lac) and Cre3.9 ratios were slightly underestimated at higher accelerations. Glx ratio on the other hand was slightly reduced at 8x while slightly increased at 12x as shown in Fig. 2.Conclusion
Due to the higher incoherence level of sampling patterns based on rosette trajectories, rosette based spectroscopic imaging sequence has the potential for highly accelerated acquisitions. A rosette based 4D J-resolved Spectroscopic Imaging sequence was implemented and the feasibly of 12x acceleration is demonstrated in this work. However, further optimization and validation using a larger cohort of in-vivo datasets is needed.Acknowledgements
Authors acknowledge grants support from National Institute of Health (5R21MH125349-02 and 5R01HL135562-04).References
1. Thomas MA, Ryner LN, Mehta MP, Turski PA, Sorenson JA. Localized 2D J‐resolved H MR spectroscopy of human brain tumors in vivo. Journal of Magnetic Resonance Imaging. 1996 May;6(3):453-9.
2. Thomas MA, Ryner LN, Mehta MP, Turski PA, Sorenson JA. Localized 2D J‐resolved H MR spectroscopy of human brain tumors in vivo. Journal of Magnetic Resonance Imaging. 1996 May;6(3):453-9.
3. Ludwig C, Viant MR. Two‐dimensional J‐resolved NMR spectroscopy: review of a key methodology in the metabolomics toolbox. Phytochemical Analysis: An International Journal of Plant Chemical and Biochemical Techniques. 2010 Jan;21(1):22-32.
4. Schulte RF, Lange T, Beck J, Meier D, Boesiger P. Improved two‐dimensional J‐resolved spectroscopy. NMR in Biomedicine: An International Journal Devoted to the Development and Application of Magnetic Resonance In vivo. 2006 Apr;19(2):264-70.
5. Mansfield P. Spatial mapping of the chemical shift in NMR. Magn Reson Med 1984;1(3):370-86. 6. Posse S, Otazo R, Caprihan A, Bustillo J, Chen H, Henry PG, et al. Proton echo‐planar spectroscopic imaging of J‐coupled resonances in human brain at 3 and 4 Tesla. Magn Reson Med 2007;58(2):236-44.
7. Furuyama JK, Wilson NE, Burns BL, Nagarajan R, Margolis DJ, Thomas MA. Application of compressed sensing to multidimensional spectroscopic imaging in human prostate. Magn Reson Med 2012;67(6):1499-505.
8. Li Y, Yang R, Zhang C, Zhang J, Jia S, Zhou Z. Analysis of generalized rosette trajectory for compressed sensing MRI. Medical physics. 2015 Sep;42(9):5530-44.
9. Schirda CV, Tanase C, Boada FE. Rosette spectroscopic imaging: optimal parameters for alias‐free, high sensitivity spectroscopic imaging. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2009 Jun;29(6):1375-85.
10. Schulte RF, Wiesinger F, Fish KM, Whitt D, Hancu I. Efficient hyperpolarised 13C metabolic imaging with rosette spectroscopic imaging. InProc Int Soc Magn Reson Med 2009 (Vol. 17, p. 2433).
11. Shen X, Özen AC, Sunjar A, Ilbey S, Sawiak S, Shi R, Chiew M, Emir U. Ultra-short T2 components imaging of the whole brain using 3D dual-echo UTE MRI with rosette k-space pattern. Magnetic Resonance in Medicine. 2022 Sep 25.
12. Sarty GE. Critical sampling in ROSE scanning. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2000 Jul;44(1):129-36.
13. Joy A, Paul JS. Multichannel compressed sensing MR image reconstruction using statistically optimized nonlinear diffusion. Magn Reson Med 2017;78(2):754-62.
14. Joy A, Paul JS. A mixed-order nonlinear diffusion compressed sensing MR image reconstruction. Magn Reson Med 2018;80(5):2215-22.
15. Joy A, Jacob M, Paul JS. Compressed sensing MRI using an interpolation‐free nonlinear diffusion model. Magn Reson Med 2021;85(3):1681-96.
16. Joy A, Jacob M, Paul JS. Directionality guided non linear diffusion compressed sensing MR image reconstruction. Magn Reson Med 2019;82(6):2326-42.
17. Fessler JA, Sutton BP. Nonuniform fast Fourier transforms using min-max interpolation. IEEE transactions on signal processing. 2003 Jan 22;51(2):560-74.
18. Schulte RF, Boesiger P. ProFit: two‐dimensional prior‐knowledge fitting of J‐resolved spectra. NMR in Biomedicine: An International Journal Devoted to the Development and Application of Magnetic Resonance In vivo. 2006 Apr;19(2):255-63.
Figures