4650
Validation of a deep-learning-based method for accelerating susceptibility-weighted imaging in clinical subjects1Radiology, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China, 2The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
Synopsis
Keywords: Data Acquisition, Machine Learning/Artificial Intelligence, deep-learning ,susceptibility-weighted imaging,magnetic resonance imaging
In this study, we validated a deep-learning-based method for accelerating susceptibility-weighted imaging (SWI) in 31 clinical subjects. Compared to the fully sampled images, the accelerated SWI images had less noise and imaging artifacts. Although the images had decreased sharpness, the anatomical details of the lesions were mostly kept, and we had not observed false negative\positive lesions. This method could be useful for clinical situations that need timely imaging results.INTRODUCTION
Susceptibility-weighted imaging (SWI) is widely used in clinical settings to characterize hemorrhage and mineralization1-3. Nonetheless, its application might be limited by the long acquisition time, especially in patients who cannot tolerate long scanning times or are in urgent need of treatment (e.g., stroke). Recently, some deep-learning (DL) based SWI acceleration methods4, 5 have been proposed, but few have performed validation in real clinical settings.METHODS
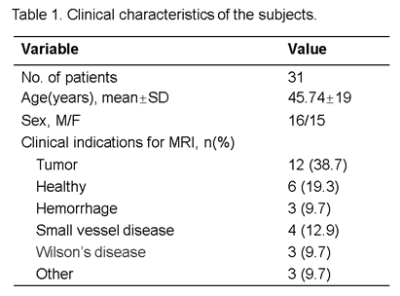
The study protocol has been approved by the ethics committee of the second affiliated hospital of Zhejiang University School of Medicine. A total of 31 subjects were consecutively enrolled in the present study. A DL accelerated SWI sequence was performed on each subject using a 24-channel head-neck coil on a uMR790 scanner (United Imaging, Shanghai, China). The parameters for the DL-SWI acquisition were TR=28.2ms, TE=20ms, Flip anlge=15°, matrix=336*292, voxel size=0.685*0.685*2mm3, acceleration factor=5. As a reference, the routine parallel imaging (PI) accelerated (AF=2) SWI scan with otherwise identical scanning was also performed. The acquisition times for both DL and PI accelerated SWI sequences were 1min46s and 4min45s, respectively. Two radiological technicians, XW (10 years of experience) and SX (9 years of experience) independently performed the image quality assessments on all images by a double-blinded procedure. Firstly, the observer evaluated the following image features: level of noise, lesion conspicuity (when present), anatomic details (brain structures, tissue boundaries), artifacts (motion and parallel imaging artifacts), and overall image quality. The scoring was from 1 to 5 for each index, representing quality from poor to excellent. For subjects with microbleeds, the observers also counted the total number of microbleeds. Secondly, both SWI image sets were put side-by-side and the observers compared the two images in each subject. With scores 1 to 5, the DL-accelerated SWI images were rated as worse (1-2), equal (3), or better than the PI SWI counterparts (4-5). The inter-rater correlation coefficient was used to assess the consistency between the two observers. The Wilcoxon signed-rank tests and student’s t-tests were used to test the difference in ordinal and continuous variables, respectively. A difference student t-test was used to investigate the noninferiority of the DL-accelerated images (noninferiority margin, -0.5).RESULTS
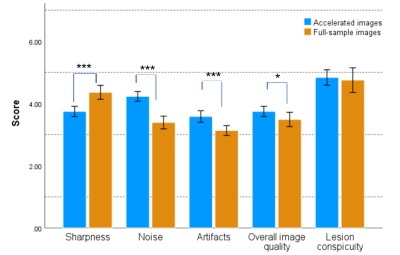
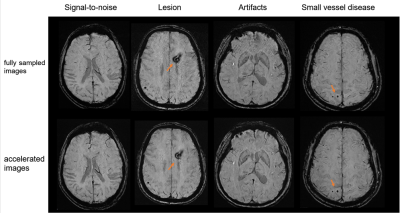
The demographics can be seen in Table 1. Compared to the fully sampled images, DL-accelerated images had significantly higher scores in terms of noise, artifacts, and overall image quality score (p < 0.001, p < 0.001, p = 0.046, respectively). The fully sampled images have higher scores than the DL-accelerated images in terms of the sharpness of the image (p < 0.001). There was no significant difference between fully sampled and accelerate images in terms of lesion conspicuity (p = 0.564[HP1] ). Additionally, the number of microbleeds identified on the DL-accelerated images completely matched the fully sampled image. Noninferiority testing showed that the DL-accelerated images (p=0.407; p=0.385).DISCUSSION
Based on our preliminary observations in a clinical setting, the DL accelerated SWI showed increased signal-to-noise and fewer imaging artifacts than the routine PI accelerated SWI with identical scan settings. The anatomical details of the lesions were mostly kept, and we had not observed any false-negative or false-positive lesions. The potential caveat of this DL accelerated SWI method is the decreased sharpness at the current resolution setting (i.e. 0.685*0.685*2mm3). As the DL reconstruction effectively filters out the highest frequency component collected, due to its Poison Disc style sampling design with more sparse sampling at the outer k-space part, the filtered spatial frequency components at the current resolution setting are those most sensitive to the human eye. This situation can be effectively improved by scanning with higher resolution such as 0.5x0.5x2mm3, which will be the interest of our future study. However, we found that the current reduction in sharpness did not interfere with clinical diagnosis, but with the benefits of greatly improved patient comfort and scan efficiency due to the significantly reduced scan times.CONCLUSION
With significantly reduced scanning time, less imaging artifacts, and mostly kept anatomical details, SWI imaging with DL acceleration could be useful for clinical situations that need timely imaging results.Acknowledgements
No acknowledgement found.References
1. Thomas B, Somasundaram S, Thamburaj K, et al. Clinical applications of susceptibility weighted MR imaging of the brain - a pictorial review. Neuroradiology 2008;50:105-116.
2. Halefoglu AM, Yousem DM. Susceptibility weighted imaging: Clinical applications and future directions. World J Radiol 2018;10:30-45.
3. Cheng AL, Batool S, McCreary CR, et al. Susceptibility-weighted imaging is more reliable than T2*-weighted gradient-recalled echo MRI for detecting microbleeds. Stroke 2013;44:2782-2786.
4. Duan C, Xiong Y, Cheng K, et al. Accelerating susceptibility-weighted imaging with deep learning by complex-valued convolutional neural network (ComplexNet): validation in clinical brain imaging. Eur Radiol 2022;32:5679-5687.
5. Kim EH, Choi MH, Lee YJ, Han D, Mostapha M, Nickel D. Deep learning-accelerated T2-weighted imaging of the prostate: Impact of further acceleration with lower spatial resolution on image quality. Eur J Radiol 2021;145:110012.
Figures