4648
Development of a self-supervised machine learning algorithm for automatic MRI sequence-type classification1Asan Medical Center, Seoul, Korea, Republic of, 2University of Ulsan College of Medicine, Seoul, Korea, Republic of, 3Ajou University Hospital, Suwon, Korea, Republic of
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence, Sequence; Classification
For automatic sequence-type classification of brain MRI, we developed the self-supervised machine learning (ML) algorithm, named ImageSort-net, using a rule-based labeling system based on metadata of Digital Imaging and Communications in Medicine (DICOM) image files. Our rule-based labeling system and ImageSort-net showed high classification performance to predict brain MRI sequence type. ImageSort-net showed reliable performance by appending a new dataset to an existing dataset and without human labeling of the whole dataset. This result indicates that sustainable self-learning ML algorithms using the rule-based virtual label in the new datasets are feasible.Background
Brain magnetic resonance imaging (MRI) is the most common imaging modality with various sequences in the stroke research field1 and it is very important to sort the brain MRI according to the specific sequence. However, medical imaging data has not been fully standardized, and the metadata of Digital Imaging and Communications in Medicine (DICOM) image files often differ across sites, vendors, and acquisition protocols. Due to the variability of DICOM metadata, it is very time-consuming to classify the brain MRI sequences according to the standardized sequence types2. Therefore, we aimed to develop the self-supervised machine learning (ML) algorithm for sequence-type classification of brain MRI using a supervisory signal from DICOM metadata (i.e., rule-based virtual labels).Methods
Two sources of datasets, i.e., the hospital dataset and the multicenter trial dataset were used in this study. A total of 1,787 brain MRI data was constructed from various hospitals and devices (1,537 and 256 in the hospital dataset and the multicenter trial dataset, respectively). To generate human expert labeling (i.e., ground truth), we manually classified and labeled eight MRI sequences that were commonly used for stroke evaluation, including T1, T2, Scout, Diffusion, MR angiography (MRA), Fluid-attenuated inversion recovery (FLAIR), susceptibility/gradient images (Susgre), and Perfusion, based on DICOM metadata attributes.
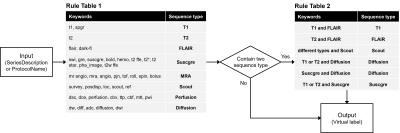
For developing the self-supervised ML algorithm, we developed a rule-based labeling system which is composed of two steps to generate virtual labels which can simulate and replace human expert labels (Figure 1).
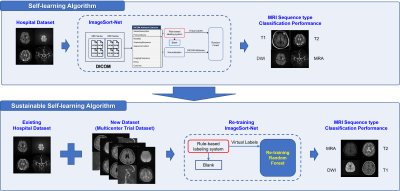
Our model, named ImageSort-net, is developed as a fully automatic MRI sequence type classifier generation architecture, composed of three steps, as follows: pre-processing step, preparation step for ML algorithm, and training step for ML algorithm using Random Forest (Figure 2). We evaluated and analyzed two models learned with human expert labels (MLhuman) and rule-based virtual labels (MLvirtual) using blank data that the rule-based labeling system failed to infer from the hospital dataset as a test set.
Additionally, we performed an experiment to add the multicenter trial dataset into the hospital dataset and train them as a whole dataset. The re-training using the whole dataset was also performed by following the three steps (Figure 2) and was no need for human labeling. The performance of the ML algorithm (MLvirtual) was performed on the whole dataset.
The performances of rule-based labeling, MLvirtual, and MLhuman algorithms in classifying the MRI sequences were evaluated. For performance measurements, we used the human expert labels of sequence types as reference standards. The F1-score, precision, recall, and accuracy were used for evaluating the classification performance.
Results
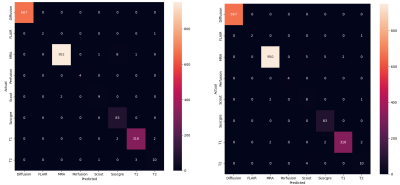
In both the hospital dataset and the multicenter trial dataset, the MRI sequence type classification performance of the rule-based labeling system which does not use machine learning was excellent with accuracy ranging from 98.6% to 99.5% (median 99.1%) and F1-score ranging from 98.0% to 98.5 % (median 98.3%).In the hospital dataset, the overall classification accuracy of MLvirtual algorithm was good as the MLhuman algorithm (98.7% and 99.0%, respectively). In each sequence type, the classification performances of MLvirtual (F1 score, 57% to 100%) were as good as the MLhuman (F1 score, 74% to 100%). Notably, the performances of MLvirtual for scout and T2 were relatively low, which might be attributed to the relatively low performance of the rule-based labeling system. Figure 3 shows the confusion matrix associated with the performance of both ML models.After integrating the hospital dataset and multicenter trial dataset into a large whole dataset, we re-trained ML model in the same way of the MLvirtual (Figure 4). The overall accuracy in the whole dataset including new data was 98.2%.
Conclusions
Training of ML algorithm based on the rule-based virtual labels achieved high accuracy for sequence type classification of brain MRI and enabled us to build a sustainable self-learning system without human labeling.Acknowledgements
This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. NRF-2020R1F1A1048267) and by the grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI18C1216).
References
1. Suh CH, Jung SC, Kim B, Cho SJ, Woo DC, Oh WY, et al. Neuroimaging in Randomized, Multi-Center Clinical Trials of Endovascular Treatment for Acute Ischemic Stroke: A Systematic Review. Korean J Radiol. 2020;21(1):42-57.
2. Gauriau R, Bridge C, Chen L, Kitamura F, Tenenholtz NA, Kirsch JE, et al. Using DICOM metadata for radiological image series categorization: a feasibility study on large clinical brain MRI datasets. Journal of digital imaging. 2020;33(3):747-62.