4640
Highly accelerated acquisition of MP2RAGE using Compressed SENSE1Department of Radiology, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, China, 2Philips Healthcare, Beijing, China
Synopsis
Keywords: Data Acquisition, Data Acquisition, compressed sensing
The magnetization-prepared 2 rapid acquisition gradient echo (MP2RAGE) sequence provides morphological T1-weighted images and quantitative T1 maps. Compressed sensing (CS) acceleration technique has been drawing extensive attention with its high acquisition efficiency for MRI sequence. In this study, volume measurement and ROI analysis were used to assess the images obtained from CS-MP2RAGE sequence under different acceleration factors. We find that CS-MP2RAGE show high similarity in brain structure volumes measurement and ROI analysis with traditional SENSE-MP2RAGE. Those findings prove that CS-MP2RAGE can be considered as an alternative to the SENSE-MP2RAGE, though the specific acceleration factor still needs to be further studied.INTRODUCTION
Morphological T1-weighted images are necessary for MRI applications, especially as an anatomical reference in neuroimaging studies.1 MPRAGE(magnetization prepared rapid gradient echo)is a commonly used sequence to acquire 3D T1-weighted image, which has been used in the Alzheimer's Disease Neuroimaging Initiative (ADNI) with high resolution. 2-3 However, the inhomogeneity of the transmit B1 + and receive B1 – fields could affect the image quality in high magnetic fields. As an extension of the MPRAGE sequence,The magnetization-prepared 2 rapid acquisition gradient echo (MP2RAGE) sequence could provide superior grey-white matter contrast T1-weighted images and quantitative T1 maps. 4 A typical MP2RAGE scan with SENSE /GRAPPA usually takes a long time.In recent years, compressed sensing (CS) acceleration technique has been drawing extensive attention with its high acquisition efficiency for MRI sequence5. Mussard6 accelerated MP2RAGE sequence using CS, and the obtained images were considered equivalent to the gold standard GRAPPA-MP2RAGE, while shortening the acquisition time by 57%. In our research, we apply various acceleration factors to all the volunteers and aimed to quantitatively assess the images obtained from CS-MP2RAGE sequence under different acceleration factors compared to traditional SENSE-MP2RAGE.
METHODS
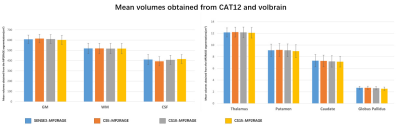
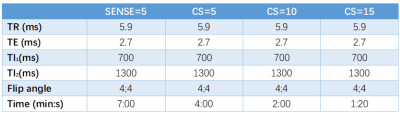
12 healthy volunteers were included who underwent MRI scan on a 3T scanner (Ingenia CX, Philips, Best, The Netherlands), using a 32-channel receive head coil. Four MP2RAGE sequences were performed for each subject and the scan parameters are listed in Table1.Firstly, volume measurements obtained from CAT12 and VolBrain were assessed. Segmentation of brain white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) was performed using Computational Anatomy Toolbox (CAT12version 1613; http://www.neuro.uni-jena.de/cat/). To evaluate subcortical structures, the online platform VolBrain software was used (https://www.volbrain.upv .es). The segmentation results obtained from all four sequence were compared using one-way ANOVA (P < 0.05 was considered significant).
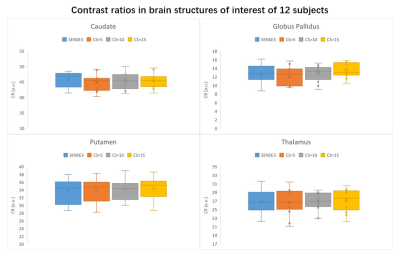
Then, the following substructures were selected for region-of-interest (ROI) analysis: putamen, thalamus, globus pallidus, caudate head, genu of corpus callosum. We chose the genu of corpus callosum as the reference WM to quantify the gray/white contrast. At these ROIs, Contrast ratios (CRs) between structures were calculated using the same equations as in Okubo et al7.
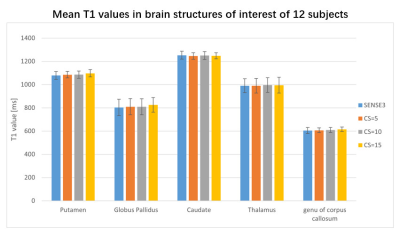
T1 values were also calculated using the ROIs as just described. The mean T1 of a structure was measured as the average of the T1 values of each voxel within the region of interest. One-way ANOVAs were also used to assess CRs 7and T1 value among all four sequences.
RESULTS
The scan time was reduced by 43%, 71% and 81% when CS acceleration factors was 5, 10 and 15 respectively, compared to 7 minutes for SENSE3, The mean and SD of the volumes of WM, GM, CSF, putamen, thalamus, globus pallidus, caudate on the 12 participants are shown in Figure1. No significant difference in volumes was observed among the four sequences. The CRs of all sequences are shown in the boxplots in Figure2. For most regions, CS-MP2RAGE images exhibited a lower CRs compared to the SENSE images, but there was no significant difference in CRs for all sequences. The measured T1 values of five ROIs are displayed in FIGURE3. Although there is a trend that higher acceleration factors lead to higher T1 values, it is not statistically significant.DISCUSSION
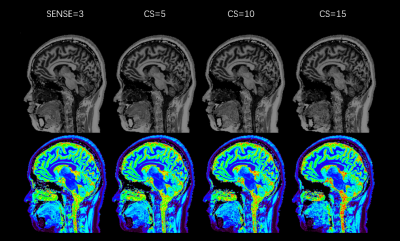
Our study is the first one to systematically evaluate the differences in volume and image quality of brain structures between the traditional SENSE-MPRAGE and CS-MP2RAGE with different acceleration parameters.Indeed, Mussard et al6 only compared the parallel imaging accelerated MP2RAGE and CS-MP2RAGE with single factor.FIGURE4. shows that the overall image quality allows distinguishing the main structures, even when CS=15. However, an increased blurring comes with higher factor and impairs the reading of precise structures such as in cerebellum or at GM–WM borders.6
Two commonly-used softwares were used here to segment the brain structures from the MP2RAGE images,CAT12 was used to segment WM, GM, CSF, and Volbrain was used to assess subcortical regions. As shown in the results, the increasing factor dose not influence both tissue segmentations and ROI analysis. The volumes of brain tissues and ROI analysis of healthy volunteers were compared between SENSE-MP2RAGE and CS-MP2RAGE. As already demonstrated, in terms of volume, CRs and T1 value, there was no statistical difference among all four sequences. In future research, we plan to add other methods to evaluate the results, such as DICE coefficient and Coefficients of variation.
CONCLUSION
CS-MP2RAGE shows high similarity in brain structure volumes measurement and ROI analysis with SENSE-MP2RAGE, even when CS=15 (the scan time is 1:20 minutes, compared to 7 minutes for the SENSE3 gold standard, a reduction of 81%). Consequently, CS-MP2RAGE can be considered as an alternative to the SENSE-MP2RAGE, though the specific acceleration factor still needs to be further studied.Acknowledgements
No acknowledgement found.References
1.Marques JP, Norris DG. How to choose the right MR sequence for your research question at 7T and above? Neuroimage. 2018; 168:119-140.
2.Mugler JP 3rd, Brookeman JR. Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE). Magn Reson Med. 1990;15(1):152-157.
3.Weiner MW, Aisen PS, Jack CR, et al. The Alzheimer’s disease neuroimaging initiative: Progress report and future plans. Alzheimers Dement. 2010; 6:202–211.
4.Marques JP, Kober T, Krueger G, van der Zwaag W, Van de Moortele PF, Gruetter R. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. NeuroImage. 2010; 49:1271–1281.
5.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007; 58:1182–1195.
6.Mussard E, Hilbert T, Forman C, Meuli R, Thiran JP, Kober T. Accelerated MP2RAGE imaging using Cartesian phyllotaxis readout and compressed sensing reconstruction. Magn Reson Med. 2020;84(4):1881-1894.
7.Okubo G, Okada T, Yamamoto A, et al. MP2RAGE for deep gray matter measurement of the brain: a comparative study with MPRAGE. J Magn Reson Imaging. 2016; 43:55–62.
Figures