4637
SuperMRF: Deep Robust Acceleration for MR Fingerprinting1Department of Electrical Engineering, University at Buffalo, State University of New York, Buffalo, NY, United States, 2Department of Biomedical Engineering, Program of Advanced Musculoskeletal Imaging (PAMI), Cleveland Clinic, Cleveland, OH, United States, 3Department of Biomedical Engineering, University at Buffalo, State University of New York, Buffalo, NY, United States, 4Paul C. Lauterbur Research Center for Biomedical Imaging, Medical AI research center, SIAT, CAS, Shenzhen, China
Synopsis
Keywords: MR Fingerprinting/Synthetic MR, Image Reconstruction
We propose a novel, deep learning-based method, “SuperMRF”, for the reconstruction of MR Fingerprinting (MRF) parametric maps that enables rapid image reconstruction. Built upon a convolutional neural network, SuperMRF uses three loss functions to incorporate additional information from the Bloch equations, estimated maps, de-aliasing, and data consistency losses. We investigate the use of SuperMRF for further acceleration of data acquisition by reducing the number of MRF time frames. Our results demonstrate that proposed SuperMRF is robust to noise and can achieve a 20x reduction in acquired MRF time frames. Tissue property maps can be reconstructed in less than one second.Introduction
Magnetic resonance fingerprinting (MRF)[1] is an acquisition scheme that adopts a variable schedule of radiofrequency excitations and delays to generate unique signal evolutions (signatures) for tissues of differing types. With MRF, multiple quantitative tissue parameter maps can be simultaneously obtained, which significantly reduces total scan time. However, obtaining accurate MRF signatures for different tissues can still require a relatively long scan time, particularly for an inner product-based pattern-matching reconstruction approach. Iterative low-rank MRF reconstruction can enable decreased acquisition time while still providing improved accuracy and precision. Nevertheless, as multiple iterations between the image domain and k-space sampled data are required, the computation is a burden. Thus, such reconstructions are generally performed offline after the MRI exam. As such, there is a need for rapid, robust reconstruction methods that can generate tissue property maps quickly, which would enable inline reconstruction at the time of the MRI exam. Deep learning technology such as DRONE[2] was introduced to train on a simulated sparse dataset, bypassing the limits imposed by the exponentially growing dictionary. However, the robustness of that method is limited. Here we propose a novel superfast and robust method for the reconstruction of MRF that incorporates spatial and temporal information as well as Bloch Equation relations via deep learning named SuperMRF. We investigate the acceleration by SuperMRF in the number of time frames in addition to the same k-space undersampling factor without compromising accuracy. We also investigate the robustness of SuperMRF to noise.Methods
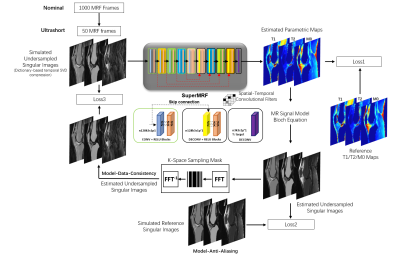
SuperMRF is a deep multi-channel framework that estimates desired parametric maps directly from undersampled weighted images. Residual learning[3] and patch-based training/padding[4, 5] are chosen to promote both accuracy and robustness. Three loss terms are used to train SuperMRF, as shown in Figure 1: Loss1 is for the direct estimated parametric maps, which is the traditional loss. The other two losses are cyclic losses. Loss2 is for de-aliasing, and Loss3 is for data consistency. Bloch Equation relationships are embedded in the network during the learning process as part of Loss2 and Loss 3, as the parametric maps estimated via SuperMRF are used in conjunction with Bloch simulation to generate MR signal evolutions. For acceleration, the first 50 undersampled image frames are used. Temporal compression into a low-rank subspace via singular value decomposition is employed to obtain 3 singular images that have reduced undersampling artifacts than the original image frames.3D Cartesian MRF Sequence
The MRF sequence used a 3D Cartesian trajectory with linear readout in kx and a pseudorandom sampling in ky-kz with Gaussian density weighting at the center of k-space. The MRF acquisition consisted of 1000 image frames that were acquired with fast imaging with steady-state free precession, 4π spoiler gradient, and each frame was acquired at a specified acceleration factor (R=15 in this work). Multiple readouts at different ky-kz sample points were achieved by repeating the 1000 frame acquisition after a time that is sufficient for approximately full longitudinal magnetization recovery (4s assumed in this work). For the MRF acquisition itself, variable flip angles between 5 to 15 degrees, constant repetition time (TR) of 10 ms, and constant echo time (TE) of 5 ms were used. An inversion pulse was applied prior to frames 1 and 501 with an inversion time of 21 ms. T2 preparation pulses were applied prior to frames 101, 201, 301, and 401 with echo times of 20, 40, 60, and 80 ms, respectively, and with the same pattern again at 601, 701, 801, and 901. Simulation experiments were performed using in vivo T1, T2, and M0 maps acquired with conventional methods (inversion recovery, T2-prepared gradient echo). Subjects provided written informed consent as part of an IRB-approved protocol.Results and Discussion
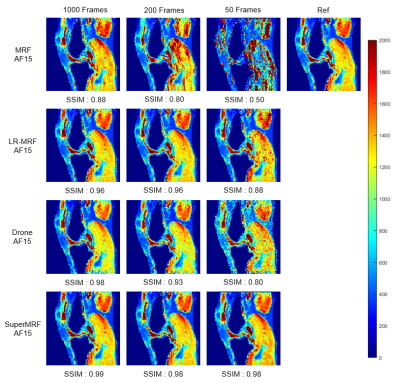
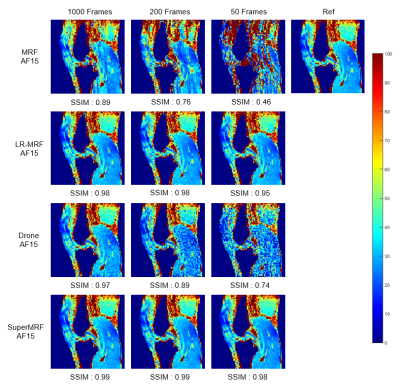
Frame reduction testFigure 2 and Figure 3 show Conventional MRF, low-rank MRF (LR-MRF), SuperMRF, and DRONE performance for T1 and T2 mapping with frames reduced from 1000 to 50. Tissue property maps reconstructed by LR-MRF and SuperMRF are less sensitive to frame reduction and can maintain a good reconstruction quality. The structural similarity index (SSIM)[6] was calculated with the 1000 frames without k-space reduction as the reference. The training required 10 hours, and reconstruction took 1 second for one complete dataset. This rapid reconstruction time can be contrasted with that required of state-of-the-art iterative, low-rank reconstruction implementations that can take hours to complete for 3D MRF datasets. Compared with the existing DL method for MRF[7], SuperMRF utilizes neighborhood information, which allows even higher reduction (both in k and frame space) and more robust reconstruction than DRONE.
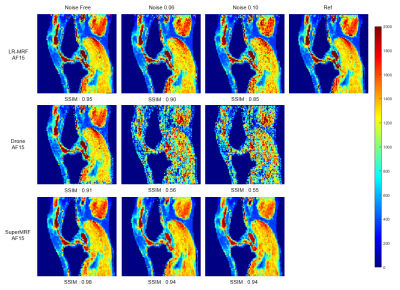
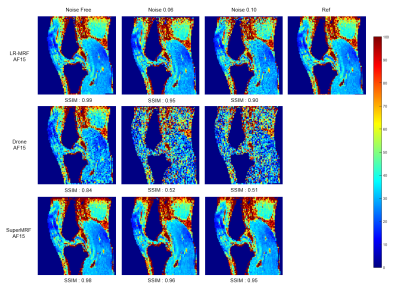
Noise robustness test
Figure 4 and Figure 5 show the noise robustness test for Conventional MRF, LR-MRF, SuperMRF, and DRONE. The parametric maps estimated by SuperMRF were close to the ground truth T1/ T2 maps, even with increasing noise.
Conclusion
We demonstrated the feasibility of a superfast MRF reconstruction technique with only 50 undersampled frames using controlled simulation experiments with realistic anatomy. Although only a single MRF sequence was used in this study, SuperMRF is expected to be complementary to other, potentially more optimized data acquisition schemes. Future work will explore a larger-scale prospective in vivo evaluation of SuperMRF to benefit a wide range of clinical and scientific studies that require superfast MRF.Acknowledgements
This work was funded in part by the following sources: NIH/NIAMS T32AR007505, NIH/NIA K25AG070321. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.References
[1] Ma, D., et al., Magnetic resonance fingerprinting. 2013. 495(7440): p. 187-192.
[2] Cohen, O., B. Zhu, and M.S.J.M.r.i.m. Rosen, MR fingerprinting deep reconstruction network (DRONE). 2018. 80(3): p. 885-894.
[3] He, K., et al. Deep residual learning for image recognition. in Proceedings of the IEEE conference on computer vision and pattern recognition. 2016.
[4] Kim, J., J. Kwon Lee, and K. Mu Lee. Accurate image super-resolution using very deep convolutional networks. in Proceedings of the IEEE conference on computer vision and pattern recognition. 2016.
[5] Dong, C., et al., Image super-resolution using deep convolutional networks. IEEE transactions on pattern analysis and machine intelligence, 2015. 38(2): p. 295-307.
[6] Wang, Z., et al., Image quality assessment: from error visibility to structural similarity. IEEE transactions on image processing, 2004. 13(4): p. 600-612.
[7] Cohen, O., B. Zhu, and M.S. Rosen, MR fingerprinting deep reconstruction network (DRONE). Magnetic resonance in medicine, 2018. 80(3): p. 885-894.
Figures