4633
Standardization of Containerized “MRD Apps” for Reproducible and Deployable Research1Siemens Medical Solutions USA, Inc., Chicago, IL, United States, 2Boston Children's Hospital, Boston, MA, United States, 3Karolinska Institutet, Karolinska, Sweden, 4National Institutes of Health, Bethesda, MD, United States, 5Friedrich-Alexander Universität, Erlangen, Germany
Synopsis
Keywords: Software Tools, Reproductive
Reproducibility and translation of advancements in MR image reconstruction and analysis algorithms have been limited by incompatibilities between heterogeneous software environments. Containerization of “MRD Apps” packages software together with all dependent libraries into a single ready-to-use container. Standardization of the container format enables broad inter-compatibility and source code examples are provided for Python, MATLAB, and C++ through Gadgetron. A neural network was implemented in the MRD Apps format using Python and run directly on the scanner with vendor-provided integration. MRD Apps can be published alongside manuscripts as reference implementations or used to streamline image reconstruction and analysis challenge contests.Introduction
Advancements in MR image reconstruction and analysis algorithms have resulted in significant improvements in acquisition time, image quality, and quantitative imaging. However, these advancements are often difficult to reproduce due to increasingly complex algorithms or neural networks whose performance is highly dependent on training data. Algorithms are also often difficult to share between researchers or deploy on clinical scanners due to incompatibilities between the heterogeneous software development environments used by researchers and vendors alike.The increasing popularity of open-source software is partially aimed at addressing these issues, but there remain significant challenges with incompatible data formats, synchronizing dependent libraries, or even compiling code due to poor documentation. Open distribution of source code may also not be possible due to licensing restrictions, intellectual property considerations, or commercial interests. Standardized and common interfaces to these applications alongside open-source applications can still be highly beneficial for reproducible science.
Containerization is a popular approach to packaging application software along with all dependent libraries together with a light-weight operating system into a single container. These can be easily shared and deployed on various systems without additional configuration, providing a reproducible, easy to use way of packaging MR applications. Containerization has been used for MR in Gadgetron (1) and challenges (2), however the lack of standardization has limited their broader adoption. We propose to develop a standardized containerization interface with wide-ranging applicability and support for common development environments.
Methods
The ISMRM Raw Data (MRD, previously known as ISMRMRD) format is an open-source, vendor neutral description of raw k-space, image, and waveform data primitives along with associated acquisition parameter metadata (3). Converters are available from multiple vendor proprietary raw data formats (4-7) and MRD images can be converted from standard DICOM images (8) or created directly in MATLAB or Python using provided libraries (3).An extension of the MRD format provides standardized serialization and deserialization of data primitives along with a client/server-based communications protocol created in Gadgetron (3,9). Data analysis occurs on the server side, which can be implemented in any programming language or framework that supports the MRD communications format, including Gadgetron, Python (8,10), and MATLAB (11,12). Servers were containerized into “MRD Applications” using Docker (13) to enable easy deployment in a reproducible manner.
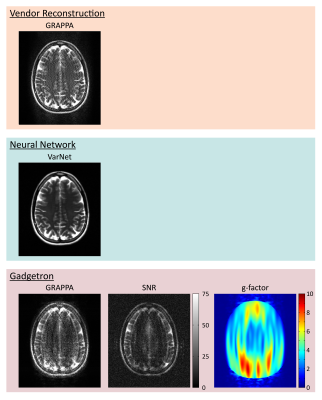
Various algorithms for image reconstruction and processing implemented in various programming frameworks were packaged as MRD Apps (Fig. 1). A variational neural network (VarNet) (14) for reconstruction of the highly accelerated fastMRI dataset (15) was adapted in Python to use the MRD App format and bundled into a Docker container (16). Source code is available online at https://github.com/kspaceKelvin/VarNet-MRD-App. The VarNet MRD App was integrated for inline deployment on a 1.5T scanner (MAGNETOM Sola, Siemens Healthcare, Erlangen, Germany) using the FIRE prototype. A T2-weighted turbo spin echo sequence with R=8 parallel acceleration was prospectively acquired in a single volunteer with both vendor-provided conventional GRAPPA parallel imaging reconstruction and inline FIRE VarNet reconstruction. MRD data from this acquisition was also reconstructed in a MRD App-compatible Gadgetron container with SNR-scaled reconstruction and g-factor estimation (17).
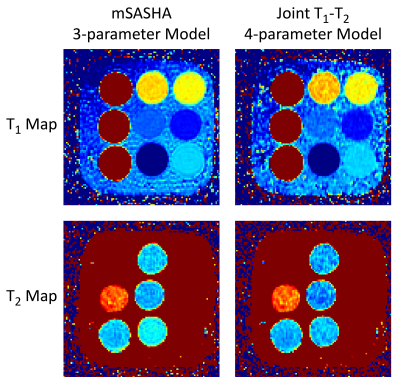
MRD Apps can also be used for image processing applications without k-space data. mSASHA is a multi-parametric cardiac T1 and T2 mapping sequence, with high precision and accuracy enabled by a 3-parameter fitting model (18). An MRD App of mSASHA mapping was implemented in MATLAB (R2022a, The MathWorks, Natick, USA) with both 3-parameter and conventional 4-parameter fitting (19). Source code is available online at: https://github.com/kspaceKelvin/mSASHA-MRD-App. Validation data from a NiCl2-doped agarose phantom (T1MES, Resonance Health, Perth, Australia) was converted to MRD images and T1 and T2 maps were calculated using both 3-parameter and 4-parameter models.
Results
The VarNet MRD App was successfully deployed inline on the scanner with significantly improved image quality compared to reference parallel imaging reconstruction (Fig. 2). Reconstruction of the same dataset using Gadgetron revealed an average whole-brain SNR of 18 with g-factors exceeding 9 in some regions. mSASHA T1 and T2 maps had an average reduction of 38% and 13% in standard deviation with 3-parameter compared to 4-parameter fitting (Fig. 3).Conclusions
MRD Apps are an extension of the widely used MRD data format, using standardized containerization to enable the reproducible deployment of algorithms across a variety of platforms including on-scanner integration. Source code examples for Python, MATLAB, and C++ through Gadgetron provide accessible starting points for most major software development platforms. Future extensions for spectroscopy could further broaden support for more MR applications. MRD Apps published alongside manuscripts can provide an easily accessible reference implementation that may facilitate adoption by others. This may also be beneficial for reconstruction or analysis challenges, where algorithms submitted as MRD Apps can be easily reused in future projects without additional effort. With vendor-provided inline integration, MRD also delivers a streamlined path for translation of research applications directly on to the scanner.Acknowledgements
No acknowledgement found.References
1. Gadgetron. Gadgetron Docker Hub. https://hub.docker.com/u/gadgetron. Accessed 9 November 2022.
2. Learn2Reg-Challenge. Learn2Reg – Grand Challenge https://learn2reg.grand-challenge.org. Accessed 9 November 2022
3. ISMRMRD Group. ISMRM Raw Data Format. https://github.com/ismrmrd/ismrmrd. Accessed 9 November 2022
4. ISMRMRD Group. Siemens ISMRMRD Converter. https://github.com/ismrmrd/siemens_to_ismrmrd. Accessed 9 November 2022
5. ISMRMRD Group. GE ISMRMRD Converter. https://github.com/ismrmrd/ge_to_ismrmrd. Accessed 9 November 2022
6. ISMRMRD Group. Philips ISMRMRD Converter. https://github.com/ismrmrd/philips_to_ismrmrd. Accessed 9 November 2022
7. ISMRMRD Group. Bruker ISMRMRD Converter. https://github.com/ismrmrd/bruker_to_ismrmrd. Accessed 9 November 2022
8. Chow K. Example client/server example for the streaming ISMRM Raw Data protocol. https://github.com/kspaceKelvin/python-ismrmrd-server. Accessed 9 November 2022
9. Gadgetron. Gadgetron - Medical Imaging Reconstruction Framework. https://github.com/gadgetron/gadgetron. Accessed 9 November 2022
10. Knudson K. Example client/server example for the streaming ISMRM Raw Data protocol. https://github.com/kristofferknudsen/gadgetron-python-ismrmrd-server. Accessed 9 November 2022
11. Fyrdahl A. An ISMRMRD-compatible image reconstruction server written in MATLAB. https://github.com/fyrdahl/matlab-ismrmrd-server. Accessed 9 November 2022
12. Chow K. An ISMRMRD-compatible image reconstruction server written in MATLAB. https://github.com/kspaceKelvin/matlab-ismrmrd-server. Accessed 9 November 2022
13. Docker. https://www.docker.com. Accessed 9 November 2022
14. Sriram A et al. End-to-End Variational Networks for Accelerated MRI Reconstruction. MICCAI 2020. doi:10.1007/978-3-030-59713-9_7.
15. Zbontar J et al. fastMRI: An Open Dataset and Benchmarks for Accelerated MRI. arXiv:1811.08839.
16. Chow K. VarNet Reconstruction as an MRD App. https://github.com/kspaceKelvin/VarNet-MRD-App. Accessed 9 November 2022
17. Kellman P et al. Image Reconstruction in SNR Units: A General Method for SNR Measurement. Magn Reson Med. 2005;54:1439
18. Chow K et al. Improved accuracy and precision with three-parameter simultaneous myocardial T1 and T2 mapping using multiparametric SASHA. Magn Reson Med. 2022;87:2775.
19. Akçakaya M et al. Joint myocardial T1 and T2 mapping using a combination of saturation recovery and T2-preparation. Magn Reson Med. 2016;76:888-896.
Figures